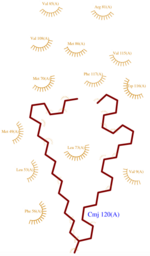
The protein AmelASP1 has been identified in the antennae from the honeybee Apis mellifera. Its primary sequence is a 144 amino acids polypeptide with a molecular weight of 13.180 kDa. AmelASP1 is part of the Pheromone Binding Protein (PBP) family. The 3D representation shown below was obtained at pH 5.5 using the nano-drops technique.
Biological function
As many other social insects, honeybees employ a large varsity of pheromones to ensure intraspecific communication in several behavioral contexts.
The social organization of the hive is strongly determined by chemical signals, also known as pheromones, that are actively produced and transmitted by the queen.
Social relevance
The diets of workers bees and queen bee are strongly different and determinate diverse behaviors.
Workers bees are fed with royal jelly for only three days after egg-laying whereas the queen bee eats royal jelly during her whole life. She controls the activity of each bees by chemical communication.
Actually, the queen bee is the only one able to produce 9-ODA - the main component of its pheromone which induces sexual or endocrine responses. This substance is sent to the workers bees which detect it through pheromone-binding proteins (PBPs). It is then transformed in 9-HDA and added in the royal jelly. This last substance is eaten by the queen. In turn, queen bee transforms 9-HDA into 9-ODA.
Thus, ASP1 is primordial to the internal pheromone’s transport cycle in the hive. By binding the component of queen bee pheromone, bees express and transmit essential behaviour within the hive. Indeed, 9-ODA is responsible, among others, of preventing workers bees’ ovarian development.
Location in the antenna and transport of pheromones
ASP1 is a protein which is only produced in the antenna of drones and workers bees. This organ constitutes one major component of the bees’ olfactory system. Cuticules structures of these antennas shelter sensillae which are a gate for pheromones. These sensillae contain neurons. Branched endings are surrounded with sensillar lymph where ASP1 captures 9-ODA and transports it to pheromone receptor in the neuron membrane. PBP’s function is to solubilize Hydrophobic odorant molecules, prevent their degradation and to transport them to reach the olfactory receptor.
Structure
Domains and family
The domain of this molecule presents a characteristic PBP-GOP domain. While this protein is composed of 144 residues the PBP domain begins at the 25th residue.
Key residues
AmelASP1 is composed of [1]
- : residues 8–25
- : residues 27–36
- : residues 42–56
- : residues 66–74
- : residues 75–77 (rarely mentioned in publications because of its tiny size)
- : residues 78–90
- : residues 96–112
has a break in the hydrogen-bonding pattern of its structure, forming tight substitute hydrogen bonds with water molecules. Thus, it results in a (at residue Ala 14) induced by a disruption in the helical conformation, due to hydrogen bonds with water molecules.[2]
Components implicated in the structure rigidity
AmelASP1 presents which are greatly enhancing its structure’s rigidity by linking four of the helices together. The six cysteins and their interval spacing are the most striking features shared by proteins belonging to the PBP family.
The is established between and through Cysteines 20 and 51. links and through Cys 47 and 98, and the connects and thanks to Cys 89 and Cys 107.
Furthermore, non covalent bonds also play an important role.
Indeed, at pH 5.5, among the numerous other, two hydrogen bonds are particularly noticeable because of their importance in the formation of the loop stabilizing . is established by Asp 66 and Leu 58 whereas is formed between Asp 60 and Ala 63.
Cavity
The dynamic structure of the protein is responsible of the ligand’s binding by adjustment of position. The successful delivery of the effector to the receptor relies on this property. The ligand binding pocket consists in a [3] formed by the helices H2, H3, H4, H5 and H6 arranged in a globular shape which leads to a clear separation of the ligand from the Polar environment.
The top of the cavity is not closed and can establish contacts with the solvent. The cavity is prone to accept ligand such as 9-ODA because of its specific composition. Indeed, cavity's components are mainly [4].They consequently interact with the ligand's Hydrophobic carbon chain and are localized on the internal face of the helix.Thus, it implies that these residues respect a regular distance pattern in the primary structure of the AmelASP1.
pH influence
pH affects the flexibility of ASP1 because it induces a different protonation state of the . Protonated residues give rise to micro-environnment changes which propagate all along the protein. Consequently, ASP1 is no longer able to interact with its ligands even if ionizable residues are distant from the cavity. [5]
In fact, depending of the pH level, Asp35 bend the C terminal domain against the cavity.
At pH 5.5, is protonated and C terminal domain isn’t bend against the cavity. While ASP1 is a monomere at acid pH, it can dimerize at neutral and basic pH.[6]
Ligands
In order to determine this Protein’s structure, several Ligand has been used at pH 5.5 because this low pH fits with its natural medium in the bee antenna.
The three ligands used to characterize and purify AmelASP1 are :
- also known as (20s)-20-Methyldotetracontane, is a serendipitous ligand. This term signify that the purification of this molecule was completely fortuitous. It is a big unsaturated mono-methyl branched carbone chain with formula C43H88. This ligand fits in the Hydrophobic cavity of AmelASP1 thanks to several interactions with
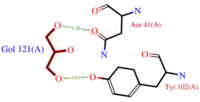
- (C3H8O3) also known as GOL, is a ligand used for cryoprotection during the purification process of the protein. It is supposedly helping the main ligand to reach its binding site.
To do so, GOL links to
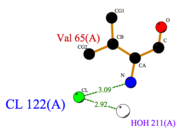
- facilitates the binding of other ligands to the protein. Its abundance around ASP1 varies according to changing pH conditions. At pH 5.5, is the only amino acid able to fix a chloride ion.
However, in natural conditions, binding ligands are the pheromones secreted by the queen such as 9-ODA.
Related structures
The structures shown below representing AmelASP1 with variable ligands and pH, emphasize and illustrate the binding versatility of Pheromones Binding Proteins.
The same PBP and their ligands can be crystalized either in apo (without ligand) or holo states (with ligand).
- In the apo state, the C terminus part of the protein obstructs the binding site.
- 3cdn The same protein in apo form soaked at pH 4.0
- 2h8v The same protein in apo form at pH 5.5
- 3cz2 The same protein in apo form at pH 7.0
- In the holo state, depending of the ligand, the volume of the cavity changes. Likewise, presence of ligand and favorable pH, induce C terminus conformation changes.
- 3fe8 The same protein in complex with a serendipitous ligand soaked at pH 4.0
- 3fe9 The same protein in complex with a serendipitous ligand soaked at pH 7.0
- 3bfa The same protein in complex with the QMP at pH 5.5
- 3bfb The same protein in complex with the 9-ODA at pH 5.5
- 3bfh The same protein in complex with the HDOA at pH 5.5
- 3bjh The same protein in complex with the nBBSA at pH 5.5
- 3cyz The same protein in complex with the 9-ODA at pH 7.0
- 3cz0 The same protein in complex with the QMP at pH 7.0
- 3cz1 The same protein in complex with the nBBSA at pH 7.0
- 3cab The same protein in complex with the nBBSA soaked at pH 7.0