This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2gk1
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:2gk1.gif|left|200px]] | + | [[Image:2gk1.gif|left|200px]] |
| - | + | ||
| - | '''X-ray crystal structure of NGT-bound HexA''' | + | {{Structure |
| + | |PDB= 2gk1 |SIZE=350|CAPTION= <scene name='initialview01'>2gk1</scene>, resolution 3.25Å | ||
| + | |SITE= | ||
| + | |LIGAND= <scene name='pdbligand=NGT:3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL'>NGT</scene> | ||
| + | |ACTIVITY= [http://en.wikipedia.org/wiki/Beta-N-acetylhexosaminidase Beta-N-acetylhexosaminidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.2.1.52 3.2.1.52] | ||
| + | |GENE= | ||
| + | }} | ||
| + | |||
| + | '''X-ray crystal structure of NGT-bound HexA''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 10: | Line 19: | ||
==About this Structure== | ==About this Structure== | ||
| - | 2GK1 is a [ | + | 2GK1 is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2GK1 OCA]. |
==Reference== | ==Reference== | ||
| - | Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis., Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN, J Mol Biol. 2006 Jun 16;359(4):913-29. Epub 2006 Apr 27. PMID:[http:// | + | Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis., Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN, J Mol Biol. 2006 Jun 16;359(4):913-29. Epub 2006 Apr 27. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/16698036 16698036] |
[[Category: Beta-N-acetylhexosaminidase]] | [[Category: Beta-N-acetylhexosaminidase]] | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| Line 24: | Line 33: | ||
[[Category: Withers, S G.]] | [[Category: Withers, S G.]] | ||
[[Category: NGT]] | [[Category: NGT]] | ||
| - | [[Category: beta-hexoasaminidase | + | [[Category: beta-hexoasaminidase some]] |
[[Category: glycosidase]] | [[Category: glycosidase]] | ||
[[Category: gm2 gangliodosis]] | [[Category: gm2 gangliodosis]] | ||
| Line 31: | Line 40: | ||
[[Category: tay-sachs disease]] | [[Category: tay-sachs disease]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 17:05:36 2008'' |
Revision as of 15:05, 20 March 2008
| |||||||
| , resolution 3.25Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Activity: | Beta-N-acetylhexosaminidase, with EC number 3.2.1.52 | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
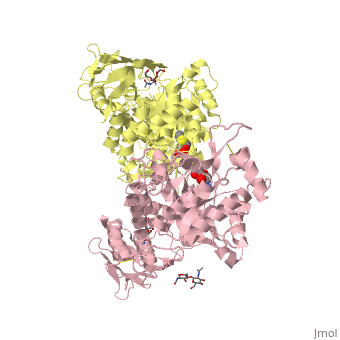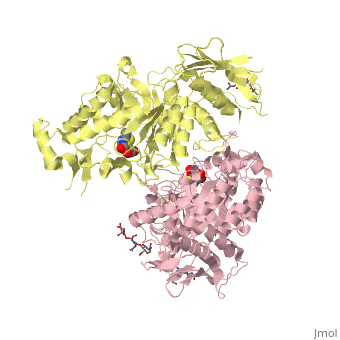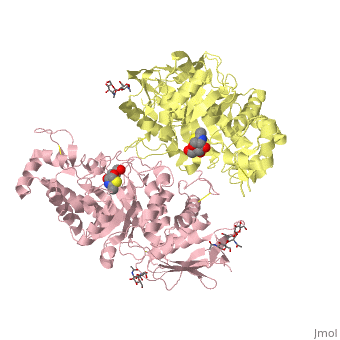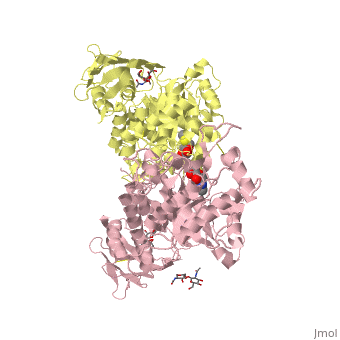
X-ray crystal structure of NGT-bound HexA
Contents |
Overview
Lysosomal beta-hexosaminidase A (Hex A) is essential for the degradation of GM2 gangliosides in the central and peripheral nervous system. Accumulation of GM2 leads to severely debilitating neurodegeneration associated with Tay-Sachs disease (TSD), Sandoff disease (SD) and AB variant. Here, we present the X-ray crystallographic structure of Hex A to 2.8 A resolution and the structure of Hex A in complex with NAG-thiazoline, (NGT) to 3.25 A resolution. NGT, a mechanism-based inhibitor, has been shown to act as a chemical chaperone that, to some extent, prevents misfolding of a Hex A mutant associated with adult onset Tay Sachs disease and, as a result, increases the residual activity of Hex A to a level above the critical threshold for disease. The crystal structure of Hex A reveals an alphabeta heterodimer, with each subunit having a functional active site. Only the alpha-subunit active site can hydrolyze GM2 gangliosides due to a flexible loop structure that is removed post-translationally from beta, and to the presence of alphaAsn423 and alphaArg424. The loop structure is involved in binding the GM2 activator protein, while alphaArg424 is critical for binding the carboxylate group of the N-acetyl-neuraminic acid residue of GM2. The beta-subunit lacks these key residues and has betaAsp452 and betaLeu453 in their place; the beta-subunit therefore cleaves only neutral substrates efficiently. Mutations in the alpha-subunit, associated with TSD, and those in the beta-subunit, associated with SD are discussed. The effect of NGT binding in the active site of a mutant Hex A and its effect on protein function is discussed.
Disease
Known diseases associated with this structure: GM2-gangliosidosis, several forms OMIM:[606869], Hex A pseudodeficiency OMIM:[606869], Sandhoff disease, infantile, juvenile, and adult forms OMIM:[606873], Spinal muscular atrophy, juvenile OMIM:[606873], Tay-Sachs disease OMIM:[606869]
About this Structure
2GK1 is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
Reference
Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis., Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN, J Mol Biol. 2006 Jun 16;359(4):913-29. Epub 2006 Apr 27. PMID:16698036
Page seeded by OCA on Thu Mar 20 17:05:36 2008