We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Tachyplesin
From Proteopedia
(Difference between revisions)
m |
|||
| Line 6: | Line 6: | ||
== Structural highlights == | == Structural highlights == | ||
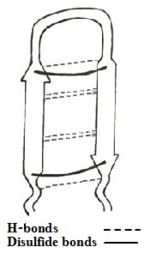
| - | Tachyplesine I is a 17-residue peptide containing six cationic residues with molecular weight 2,269 and isoelectric point (pI) of 9.93.<ref name=Chen> | + | Tachyplesine I is a 17-residue peptide containing six cationic residues with molecular weight 2,269 and isoelectric point (pI) of 9.93.<ref name=Chen>Chen, Yixin, et al. "RGD-Tachyplesin inhibits tumor growth." Cancer research 61.6 (2001): 2434-2438.</ref> |
The amino acid sequence of the TP-I is NH₂-Lys-Trp-Cys-Phe-Arg-Val-Cys-Tyr-Arg-Gly-Ile-Cys-Tyr-Arg-Arg-Cys-Arg-CONH₂. | The amino acid sequence of the TP-I is NH₂-Lys-Trp-Cys-Phe-Arg-Val-Cys-Tyr-Arg-Gly-Ile-Cys-Tyr-Arg-Arg-Cys-Arg-CONH₂. | ||
[[Image:scheme1.jpg|150px|left|thumb|<b>Figure 1: Simplefied model of Tachyplesin I.</b>]] | [[Image:scheme1.jpg|150px|left|thumb|<b>Figure 1: Simplefied model of Tachyplesin I.</b>]] | ||
| Line 52: | Line 52: | ||
The interactions between CDT and LPS may lead to a plausible disruption or fluidization of LPS structures facilitating traversal of the peptide through the LPS-outer membrane.<ref name=Saravanan>PMID:22464970</ref> | The interactions between CDT and LPS may lead to a plausible disruption or fluidization of LPS structures facilitating traversal of the peptide through the LPS-outer membrane.<ref name=Saravanan>PMID:22464970</ref> | ||
| + | |||
== Mode of action == | == Mode of action == | ||
TP-I has affinity to LPS and also has ability to permeabilize the cell membrane of pathogens. Docking model suggests strong affinity between TP-I and LPS; gained by interaction between cationic residues of TP-I with phosphate group and sachharides of LPS. Furthermore, interaction between hydrophobic residues of TP-I with acyl chains of LPS strengthens the TP-I/LPS interaction. The binding of TP-I/LPS neutralizes LPS, which is widely considered as endotoxin, and disrupts membrane function. In addition to LPS binding, footpriting analysis has revealed the binding of TP-I to DNA by interacting specifically in minor groove of DNA duplex. The interaction between TP-I and DNA is contributed by secondary structure of the peptide which contains an antiparallel beta-sheet constrained by two disulfide bridges and connected by β-turn <ref name=Yonezawa>PMID:1372516</ref>. | TP-I has affinity to LPS and also has ability to permeabilize the cell membrane of pathogens. Docking model suggests strong affinity between TP-I and LPS; gained by interaction between cationic residues of TP-I with phosphate group and sachharides of LPS. Furthermore, interaction between hydrophobic residues of TP-I with acyl chains of LPS strengthens the TP-I/LPS interaction. The binding of TP-I/LPS neutralizes LPS, which is widely considered as endotoxin, and disrupts membrane function. In addition to LPS binding, footpriting analysis has revealed the binding of TP-I to DNA by interacting specifically in minor groove of DNA duplex. The interaction between TP-I and DNA is contributed by secondary structure of the peptide which contains an antiparallel beta-sheet constrained by two disulfide bridges and connected by β-turn <ref name=Yonezawa>PMID:1372516</ref>. | ||
| + | By binding to DNA and RNA TP-1 inhibits the synthesis of macromolecules.<ref name=Hong>Hong, Jun, et al. "Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014).</ref> | ||
== Importance and relevance == | == Importance and relevance == | ||
Evidences suggest that TP-1 has ability to permeabilize the cell membranes of pathogens.<ref name=Laederach>PMID:12369825</ref>. Also, LPS and DNA being the potential biological targets of the peptide, its antimicrobial activity might be exploited. Eyeing the potential of TP-1, it has been insetred successfully in genome of ''Ornithogalum dubium'' and ''Ornithogalum thyrsoides''. These ornamentals plants were originally sensitive to soft rot erwinias (SREs) and insertion of TPI in the plants has successfully protected them without affecting their normal physiology <ref name=Lipsky>PMID:25438795</ref>. | Evidences suggest that TP-1 has ability to permeabilize the cell membranes of pathogens.<ref name=Laederach>PMID:12369825</ref>. Also, LPS and DNA being the potential biological targets of the peptide, its antimicrobial activity might be exploited. Eyeing the potential of TP-1, it has been insetred successfully in genome of ''Ornithogalum dubium'' and ''Ornithogalum thyrsoides''. These ornamentals plants were originally sensitive to soft rot erwinias (SREs) and insertion of TPI in the plants has successfully protected them without affecting their normal physiology <ref name=Lipsky>PMID:25438795</ref>. | ||
Revision as of 12:04, 6 January 2015
Introduction
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Laederach A, Andreotti AH, Fulton DB. Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry. 2002 Oct 15;41(41):12359-68. PMID:12369825
- ↑ 2.0 2.1 Chen, Yixin, et al. "RGD-Tachyplesin inhibits tumor growth." Cancer research 61.6 (2001): 2434-2438.
- ↑ Nakamura, Takanori, et al. "Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure." Journal of Biological Chemistry 263.32 (1988): 16709-16713
- ↑ Kushibiki T, Kamiya M, Aizawa T, Kumaki Y, Kikukawa T, Mizuguchi M, Demura M, Kawabata SI, Kawano K. Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim Biophys Acta. 2014 Jan 2;1844(3):527-534. doi:, 10.1016/j.bbapap.2013.12.017. PMID:24389234 doi:http://dx.doi.org/10.1016/j.bbapap.2013.12.017
- ↑ 5.0 5.1 Saravanan R, Mohanram H, Joshi M, Domadia PN, Torres J, Ruedl C, Bhattacharjya S. Structure, activity and interactions of the cysteine deleted analog of tachyplesin-1 with lipopolysaccharide micelle: Mechanistic insights into outer-membrane permeabilization and endotoxin neutralization. Biochim Biophys Acta. 2012 Mar 23;1818(7):1613-1624. PMID:22464970 doi:10.1016/j.bbamem.2012.03.015
- ↑ Yonezawa A, Kuwahara J, Fujii N, Sugiura Y. Binding of tachyplesin I to DNA revealed by footprinting analysis: significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry. 1992 Mar 24;31(11):2998-3004. PMID:1372516
- ↑ Hong, Jun, et al. "Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry." Microbiological research (2014).
- ↑ Lipsky A, Cohen A, Ion A, Yedidia I. Genetic transformation of Ornithogalum via particle bombardment and generation of Pectobacterium carotovorum-resistant plants. Plant Sci. 2014 Nov;228:150-8. doi: 10.1016/j.plantsci.2014.02.002. Epub 2014 Feb, 12. PMID:25438795 doi:http://dx.doi.org/10.1016/j.plantsci.2014.02.002
Proteopedia Page Contributors and Editors (what is this?)
Shulamit Idzikowski, Janak Raj Joshi, Michal Harel, Alexander Berchansky, Joel L. Sussman, Angel Herraez, Jaime Prilusky