We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 964
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
1f2w is a protein from the Carbonic anhydrase II (gene name CA2) sub-sub-family, which is one of the fourteen isoforms of human α carbonic anhydrases. | 1f2w is a protein from the Carbonic anhydrase II (gene name CA2) sub-sub-family, which is one of the fourteen isoforms of human α carbonic anhydrases. | ||
| - | |||
| - | Carbonic anhydrase II is located in the cytosol. | ||
This enzyme is a lyase, which is able to break C-N links, and needs its cofactor, the zinc ion, to be activated. | This enzyme is a lyase, which is able to break C-N links, and needs its cofactor, the zinc ion, to be activated. | ||
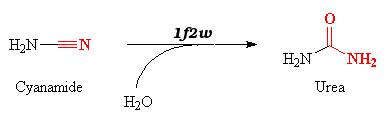
| - | The 1f2w protein catalyzes reversible hydration of cyanamide into urea. | + | Carbonic anhydrase II is located in the cytosol, and normally catalyzes the reversible hydration of CO2 into bicarbonate. |
| + | |||
| + | The 1f2w protein catalyzes reversible hydration of cyanamide into urea, which is an inhibitor of the carbonic anhydrase. | ||
==Structure== | ==Structure== | ||
| Line 23: | Line 23: | ||
- 15 <scene name='60/604483/Beta_sheets/2'>β sheets</scene> | - 15 <scene name='60/604483/Beta_sheets/2'>β sheets</scene> | ||
| - | The protein's active site is formed of <scene name='60/604483/Zinc_binding_site/2'>three histidines</scene> (residues 94,96 and 119) that can bind a zinc ion. The active site is located in an hydrophobic hole : | + | The protein's '''active site''' is formed of <scene name='60/604483/Zinc_binding_site/2'>three histidines</scene> (residues 94,96 and 119) that can bind a zinc ion. The active site is located in an '''hydrophobic hole''' : |
[[Image:125.png|400px]] | [[Image:125.png|400px]] | ||
| + | |||
| + | There is two binding sites for mercury : | ||
| + | |||
| + | - <scene name='60/604483/Hg_binding_site/1'>Hg binding site</scene>. | ||
| + | |||
| + | - <scene name='60/604483/Hgb_binding_site/1'>HgB binding site</scene>. | ||
We can see here the conservation of the residues: <jmol> | We can see here the conservation of the residues: <jmol> | ||
| Line 38: | Line 44: | ||
The most conserved residues are located in the active site's cavity. | The most conserved residues are located in the active site's cavity. | ||
| - | |||
| - | <scene name='60/604483/Hg_binding_site/1'>Hg binding site</scene>. | ||
| - | |||
| - | <scene name='60/604483/Hgb_binding_site/1'>HgB binding site</scene>. | ||
| - | |||
| - | <scene name='60/604483/Cnn_binding_site/2'>Cyanamide binding site</scene>. | ||
== Function == | == Function == | ||
| Line 51: | Line 51: | ||
[[Image:Cyanamide.png]] | [[Image:Cyanamide.png]] | ||
| + | Cyanamide is a toxic compound, and is an analog of CO2. It can thereby bind the active site of the carbonic anhydrase. | ||
| + | |||
| + | The reaction is a '''suicide inhibition''': the enzyme binds an suicide substrate (here cyanamide), and this substrate is modified by the enzyme (here into urea) and produces a reactive group that forms a '''stable inhibitor-enzyme complex'''. | ||
==Mechanism of Action== | ==Mechanism of Action== | ||
| - | The | + | The <scene name='60/604483/Cnn_binding_site/2'>Cyanamide</scene> can bind the metal ion and two threonine residues (THR 199 and 200), it is thereby adding to the coordination sphere. The cyanamid attacks the zinc ion (nucleophilic attack). Afertwards the water molecule performs a nucleophilic attack on the zinc-activated cyanamide substrate forming urea which remains bound to the metal. |
| + | |||
| + | Urea is tightly linked to the carbonic anhydrase II, acting in this way as an inhibitor. | ||
| - | The three-dimensional structure of a possible intermediate in the hydration reaction of cyanamide to urea catalyzed by human carbonic anhydrase II (hCAII) has been determined by cryocrystallographic techniques. The crystal structure shows that two different adducts are formed under the experimental conditions and that they have different occupancy in the crystal. The high occupancy form consists of a binary hCAII-cyanamide complex where the substrate has replaced the zinc-bound hydroxide anion present in the native enzyme, maintaining the tetrahedral geometry around the metal ion. The second, low-occupancy form consists of a hCAII-cyanamide-water ternary complex where the catalytic zinc ion, still being bound to cyanamide, is approached by a water molecule in a five-coordinate adduct. While the first form can be considered a nonproductive complex, the second form may represent an intermediate state of the catalyzed reaction where the water molecule is about to perform a nucleophilic attack on the zinc-activated cyanamide substrate. The structural evidence is consistent with the kinetic data previously reported about this recently described hydrolytic reaction catalyzed by hCAII, and indicates that a different mechanism with respect to that generally accepted for the physiologic carbon dioxide hydration reaction may be adopted by the enzyme, depending on the substrate chemical properties. | ||
== Disease == | == Disease == | ||
| - | [CAH2_HUMAN] Defects in CA2 are the cause of osteopetrosis autosomal recessive type 3 (OPTB3) [MIM:259730]; also known as osteopetrosis with renal tubular acidosis, carbonic anhydrase II deficiency syndrome, Guibaud-Vainsel syndrome or marble brain disease. Osteopetrosis is a rare genetic disease characterized by abnormally dense bone, due to defective resorption of immature bone. The disorder occurs in two forms: a severe autosomal recessive form occurring in utero, infancy, or childhood, and a benign autosomal dominant form occurring in adolescence or adulthood. Autosomal recessive osteopetrosis is usually associated with normal or elevated amount of non-functional osteoclasts. OPTB3 is associated with renal tubular acidosis, cerebral calcification (marble brain disease) and in some cases with mental retardation.[1] [2] [3] [4] [5] | ||
| - | + | Defects in CA2 are the cause of osteopetrosis autosomal recessive type 3 (OPTB3) [MIM:259730]; also known as osteopetrosis with renal tubular acidosis, carbonic anhydrase II deficiency syndrome, Guibaud-Vainsel syndrome or marble brain disease. Osteopetrosis is a rare genetic disease characterized by abnormally dense bone, due to defective resorption of immature bone. The disorder occurs in two forms: a severe autosomal recessive form occurring in utero, infancy, or childhood, and a benign autosomal dominant form occurring in adolescence or adulthood. Autosomal recessive osteopetrosis is usually associated with normal or elevated amount of non-functional osteoclasts. OPTB3 is associated with renal tubular acidosis, cerebral calcification (marble brain disease) and in some cases with mental retardation.[1] [2] [3] [4] [5] | |
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:31, 8 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
1f2w: Carbonic Anhydrase II
| |||||||||||