We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | ==Your Heading Here (maybe something like 'Structure')== | ||
<StructureSection load='3cww' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='3cww' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| Line 35: | Line 33: | ||
==Bradykinin== | ==Bradykinin== | ||
| - | Bradykinin is a short nonapeptide of the family of kinins. It's in response to an inflammatory envent and serves as a mediator of pain, inflammation and vasodilatation. | + | ===Bradykinin biological roles=== |
| + | Bradykinin is a short lived nonapeptide mediator of the family of kinins. It's secreted in response to an inflammatory envent and serves as a mediator of pain, inflammation and vasodilatation. | ||
| + | ===Bradykinin synthesis=== | ||
| + | it is secreted by the enzymatic action of kallikreins on kininogen precursors (?). Kallidin is a produt of the enzymatic action of kallikrein to kininogens. Then Kallidin is transformed into bradykinin | ||
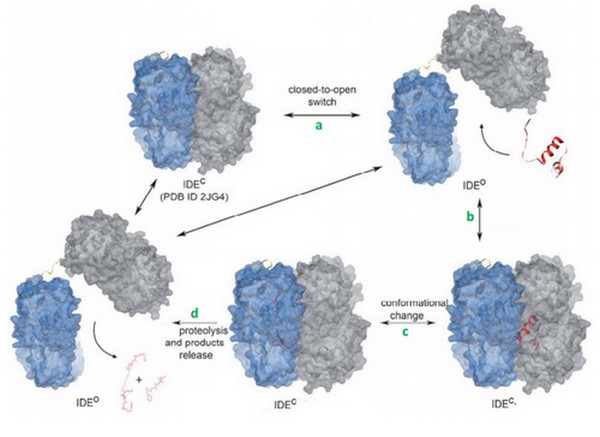
Today, we know that kinins can be degraded by IDE. | Today, we know that kinins can be degraded by IDE. | ||
Revision as of 06:49, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644