We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 951
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
== Global Structure == | == Global Structure == | ||
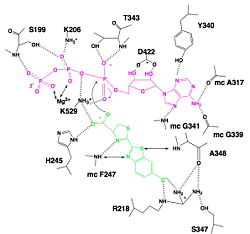
| - | Luciferase is a 62kDa protein. It contains 550 amino acids. The enzyme can be divide into two domains. On the one hand, the major portion, corresponding to the <scene name='60/604470/N_terminal_domain/1'>N-terminal</scene>. On the other hand, the small portion, corresponding to the <scene name='60/604470/C_terminal_domain/1'>C-terminal domain</scene>.Those two domains are separated by a large cleft.<ref>PMID:8805533</ref> | + | Luciferase is a 62kDa protein. It contains 550 amino acids. The enzyme can be divide into two domains. On the one hand, the major portion, corresponding to the <scene name='60/604470/N_terminal_domain/1'>N-terminal</scene>. On the other hand, the small portion, corresponding to the <scene name='60/604470/C_terminal_domain/1'>C-terminal domain</scene>.Those two domains are separated by a large cleft.<ref name =''first''>PMID:8805533</ref> |
====The C terminal domain==== | ====The C terminal domain==== | ||
| - | The amino acids of C terminal sequence is composed of contiguous residues and form a type of lid upon the N terminal domain. It contains two β sheets: the <scene name='60/604470/Beta_sheet/2'>first β sheet </scene> is composed of two short antiparallel strands and the <scene name='60/604470/Beta_sheet_2/1'> second β sheet</scene> is composed of 3 antiparallel strands, mixed with <scene name='60/604470/3_alpha_helices/1'>three α helices</scene>. Those helices are put toward the outside.It is α + β structure.<ref>PMID:8805533</ref> | + | The amino acids of C terminal sequence is composed of contiguous residues and form a type of lid upon the N terminal domain. It contains two β sheets: the <scene name='60/604470/Beta_sheet/2'>first β sheet </scene> is composed of two short antiparallel strands and the <scene name='60/604470/Beta_sheet_2/1'> second β sheet</scene> is composed of 3 antiparallel strands, mixed with <scene name='60/604470/3_alpha_helices/1'>three α helices</scene>. Those helices are put toward the outside.It is α + β structure.<ref name =''second''>PMID:8805533</ref> |
====The N terminal domain==== | ====The N terminal domain==== | ||
| - | The amino acids of C terminal sequence is composed of non contiguous residues. It falls into three subdomains (<scene name='60/604470/Domain_a/1'>the subdomain A</scene>,<scene name='60/604470/Domain_b/1'>the subdomain B</scene>, and <scene name='60/604470/Domain_c/1'>the subdomain C</scene>).It has an antiparallel <scene name='60/604470/Beta_barrel/3'>β barrel</scene>, two β sheets, which are framed by <scene name='60/604470/Alpha_helices/1'>α helices</scene>. Each of the two β sheets subdomains (A and B)are composed of 8 β strands and 6 helices. The β sheet A has <scene name='60/604470/A_beta_strands/1'>5 parallel and 3 antiparallel β strands</scene>. The β sheet B has <scene name='60/604470/A_beta_strands/1'>6 parallel and 2 antiparallel β strands</scene>. Those β sheets create a groove, closed on one end by the β barrel.<ref>PMID:8805533</ref> | + | The amino acids of C terminal sequence is composed of non contiguous residues. It falls into three subdomains (<scene name='60/604470/Domain_a/1'>the subdomain A</scene>,<scene name='60/604470/Domain_b/1'>the subdomain B</scene>, and <scene name='60/604470/Domain_c/1'>the subdomain C</scene>).It has an antiparallel <scene name='60/604470/Beta_barrel/3'>β barrel</scene>, two β sheets, which are framed by <scene name='60/604470/Alpha_helices/1'>α helices</scene>. Each of the two β sheets subdomains (A and B)are composed of 8 β strands and 6 helices. The β sheet A has <scene name='60/604470/A_beta_strands/1'>5 parallel and 3 antiparallel β strands</scene>. The β sheet B has <scene name='60/604470/A_beta_strands/1'>6 parallel and 2 antiparallel β strands</scene>. Those β sheets create a groove, closed on one end by the β barrel.<ref name =''third''>PMID:8805533</ref> |
Revision as of 11:00, 9 January 2015
| |||||||||||
References
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ 2.0 2.1 2.2 Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Photobiology
- ↑ Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Photobiology
- ↑ Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Photobiology
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ Hosseinkhani S. Molecular enigma of multicolor bioluminescence of firefly luciferase. Cell Mol Life Sci. 2011 Apr;68(7):1167-82. doi: 10.1007/s00018-010-0607-0. Epub, 2010 Dec 28. PMID:21188462 doi:http://dx.doi.org/10.1007/s00018-010-0607-0
- ↑ Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell Mol Life Sci. 2010 Feb;67(3):387-404. Epub 2009 Oct 27. PMID:19859663 doi:10.1007/s00018-009-0170-8