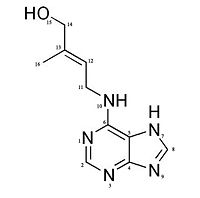
Zeatin (member of cytokinin family) Structure
Cytokinin deshydrogenase, also called CKX, is an enzyme which degrades cytokinin hormones in plants and which is encoded by the gene ZmCKX1. The one is located in Zea Maize on the chromosome 3 and belongs to a multigene family called Vanillyl-Alcohol Oxidase (VAO) flavoprotein family. The gene is more particularly expressed in the kernel of maize, mainly in the embryo since it may protect the embryo from too much cytokinin and permit the correct development. [1]
Cytokine deshydrogenase are extracellular and monomeric proteins with a molecular weight of 63kDa.[2][3] For protein production purposes, ZmCKO1 precursor protein was truncated by deletion of 18 N-terminal amino acids to produce the expected mature enzyme. [4]
The Enzyme Classification number of CKX is EC 1.5.99.12 and this indicates that the enzyme is an oxydo reductase which acts on the CH-NH group of the donor. Consequently the reaction of CKX with its cytokinin substrate is a transfer of two electrons from the cytokinin to an electron acceptor which is in the case of CKX the Flavin Adenine Nucleotide (FAD) cofactor. [2]
In some papers the denomination CKO can be found for cytokinin deshydrogenase. Indeed in the VAO flavoprotein family most of the enzyme use molecular oxygen as electron acceptor to reoxidize the FAD cofactor. That’s why the enzyme was first called Cytokinin Oxidase (CKO). CKX is an exception in the family since the enzyme uses other compounds ,such as quinone, for electron acceptor and poorly reacts with oxygen. Consequently the enzyme is now called CKX and enters the category of dehydrogenase. [1] [5]
Structure & Function
Motifs & Domains
CKX has a two main domains structure, with an (residues 33–244 and 492–534) and a (residues 245–491). [2]
The active site contains very highly conserved residues except for one residue which is located at the entrance of the active site (where a few amino acids such as Glu, Asp, Ser, Gly and other aliphatic amino acids can be found). [2]
Three residues, may have an importance in cytokinin binding and enzyme action [4]
The presence of a conserved at position 104-106 domain has been shown in the FAD binding domain which may have been highlighted in CKX structure like around position 375 and signature at the C-terminal ends. These sequences are specifically found in CKX family enzymes and their very high conservation inside the family shows that they have an important role for the enzyme functioning, such as in this case substrate recognition and electron transport. [6]
Catalytic centre
Thanks to a X-ray analysis the architecture of the catalytic centre has been determined in details. The one is made of a funnel-region on the surface of the protein and an internal cavity where we can find the flavin ring. The cavity has a volume of about 400 Angtröm and in addition to the FAD it will also be able to bind the reactive part of the substrate. This cavity has a connection to the outside thanks to a narrow pore which has an entrance containing three conserved amino acids: .
The funnel region and the cavity are also connected thanks to a pore with a 4 Angström diameter. [5]
FAD cofactor binding domain
The FAD cofactor has a covalent way of binding with the enzyme. The attachment takes place at the His105 in the GHS domain of the enzyme. The one binds the (isoalloxazine ring).
The two negative charges brought by the pyrophosphate of the FAD cofactor is compensated by the nitrogen atoms of a few amino acids such as .
Substrate binding domain
The substrate binds by following a “plug-into-socket” mode and this way of binding will cause no conformational change during binding at the active site in contrary to a lot of other dehydrogenases and oxidases. This binding mode will seal the active site. In order to have no need of movements the CKX enzyme shows a very good pre-organisation and complementary with the cytokinin substrate. There are three amino acids which play a key role in the enzyme activation: . The Asp169 residue of the enzyme is Hydrogen-bonded to the N10 atom of the substrate (various studies have shown that it is involved in the recognition of the substrate) and to the Glu288 residue. The Asp169-Glu288 pair implies that a proton is shared between the two side-chains. We can’t know the exact protonation state but the pair suppose Asp169 is an active-site base and possibly involved in proton abstraction from the substrate N10 atom. The substrate is also recognized by Glu381 which is Hydrogen-bonded to the N7 atom of the cytokinin. The others N-atoms are Hydrogen-bonded with the solvent except for N1 atom. The end of the substrate aromatic chain is located between the N10 atom of FAD, Leu492 and Asn399. These interactions allow the positioning the C11 atom of the cytokinin substrate very close to the flavin N5 atom (at about 3.0 Angström). Indeed it is this carbon atom which will be the site of the oxydative attack of the cytokinin by the FAD cofactor [5] [2]
Mechanism
The positioning of the C11 close to the N5 of the flavin suggests that electrons and protons are exchanged through a direct transfer from the substrate to the flavin. This implies a short-lived radical intermediate or the direct tranfer of a hydride anion.[5] The attack of the N5 to the C11 create a carbocation, on the cytokinin, which is stabilized by the N10–Asp 169 Hydrogen-bond interaction and the resonance effect in the oxidised imine product.
The reduced enzyme-product complex (FADH2) is reoxidised by using an organic electron acceptor, the hydroxamic acid 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-on (DIMBOA) or more precisely, the free radicals generated by the reaction of laccase and peroxydase on the DIMBOA. The reoxidation of FADH2 allows the product (adenine and 3 methyl 2 butanal) release.[5]
Regulation
The reductive part of reaction is fast, as a consequence it is the reoxidation of the FAD cofactor which constitute the rate limiting step for the catalytic action of the CKX enzyme. [4] In absence of organic electron acceptor, oxygen can be used but it is a poor electron acceptor, so this shows that the regeneration of free radicals is an important step. The coupling of CKX with laccase or peroxidase involves that the regulation of the CKX activity is highly dependent of the regulation of these two enzymes.
Different in-vivo competitive inhibitors of the CKX have been identified such as diphenylureas and derivatives such as CPPU and thidiazuron [1], which are known as cytokinin agonists [4] or Urea-type cytokinins.[2]
Another type of regulation is made by glycosylation : CKX has indeed five asparagine residues which can be glycosylated: . Protein glycosylation is particularly important since it regulates the enzymatic activity and the protein stability [2][6] and it was suggested, it has a role for enzyme localization. [1]
It has been shown that the cytokinin has a positiv feedback on the CKX activity. This indicates that the gene expression or the enzyme activity is enhanced by its substrate. It has also been shown that phyto-hormon auxin activates CKX activity and in contrary abscisic acid inhibits its activity but the relation has not been observed in the whole plant but only in certain tissues. [1]


