This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Editing User:Pauline Hanns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
Cytokine deshydrogenase are '''extracellular''' and '''monomeric''' proteins with a molecular weight of 63kDa.<ref name="Kopečnýa">PMID: 20478354 </ref><ref name="Kopečnýa2">PMID: 18571199 </ref> For protein production purposes, ZmCKO1 precursor protein was truncated by deletion of 18 N-terminal amino acids to produce the expected mature enzyme. <ref name="Kopečnýa3">PMID: 15927342 </ref> | Cytokine deshydrogenase are '''extracellular''' and '''monomeric''' proteins with a molecular weight of 63kDa.<ref name="Kopečnýa">PMID: 20478354 </ref><ref name="Kopečnýa2">PMID: 18571199 </ref> For protein production purposes, ZmCKO1 precursor protein was truncated by deletion of 18 N-terminal amino acids to produce the expected mature enzyme. <ref name="Kopečnýa3">PMID: 15927342 </ref> | ||
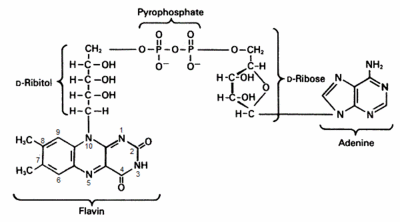
The Enzyme Classification number of CKX is EC [http://www.brenda-enzymes.info/php/result_flat.php4? 1.5.99.12] and this indicates that the enzyme is an '''oxydo reductase''' which acts on the CH-NH group of the donor. Consequently the reaction of CKX with its cytokinin substrate is a '''transfer of two electrons''' from the cytokinin to an '''electron acceptor''' which is in the case of CKX the '''F'''lavin '''A'''denine '''N'''ucleotide (FAD) cofactor. <ref name="Kopečnýa">PMID: 20478354 </ref> | The Enzyme Classification number of CKX is EC [http://www.brenda-enzymes.info/php/result_flat.php4? 1.5.99.12] and this indicates that the enzyme is an '''oxydo reductase''' which acts on the CH-NH group of the donor. Consequently the reaction of CKX with its cytokinin substrate is a '''transfer of two electrons''' from the cytokinin to an '''electron acceptor''' which is in the case of CKX the '''F'''lavin '''A'''denine '''N'''ucleotide (FAD) cofactor. <ref name="Kopečnýa">PMID: 20478354 </ref> | ||
| - | [[Image:FADESBS.gif|right| | + | [[Image:FADESBS.gif|right|400px|thumb|'''F'''lavin '''A'''denine '''D'''inucleotide]] |
In some papers the denomination '''CKO''' can be found for cytokinin deshydrogenase. Indeed in the VAO flavoprotein family most of the enzyme use '''molecular oxygen''' as electron acceptor to '''reoxidize''' the FAD cofactor. That’s why the enzyme was first called '''Cytokinin Oxidase''' (CKO). CKX is an exception in the family since the enzyme uses '''other compounds''' ,such as quinone, for electron acceptor and poorly reacts with oxygen. Consequently the enzyme is now called '''CKX''' and enters the category of '''dehydrogenase'''. <ref name="Frebortova">PMID: 19912568 </ref> <ref name="Malitoa">PMID: 15321719 </ref> | In some papers the denomination '''CKO''' can be found for cytokinin deshydrogenase. Indeed in the VAO flavoprotein family most of the enzyme use '''molecular oxygen''' as electron acceptor to '''reoxidize''' the FAD cofactor. That’s why the enzyme was first called '''Cytokinin Oxidase''' (CKO). CKX is an exception in the family since the enzyme uses '''other compounds''' ,such as quinone, for electron acceptor and poorly reacts with oxygen. Consequently the enzyme is now called '''CKX''' and enters the category of '''dehydrogenase'''. <ref name="Frebortova">PMID: 19912568 </ref> <ref name="Malitoa">PMID: 15321719 </ref> | ||
| Line 39: | Line 39: | ||
The FAD cofactor has a covalent way of binding with the enzyme. The attachment takes place at the His105 in the GHS domain of the enzyme. The one binds the <scene name='41/413140/His105_and_fad/1'> 8-methyl group of the flavin ring</scene> (isoalloxazine ring). | The FAD cofactor has a covalent way of binding with the enzyme. The attachment takes place at the His105 in the GHS domain of the enzyme. The one binds the <scene name='41/413140/His105_and_fad/1'> 8-methyl group of the flavin ring</scene> (isoalloxazine ring). | ||
| - | The two negative charges brought by the pyrophosphate of the FAD cofactor is compensated by the nitrogen atoms of a few amino acids such as <scene name='68/686754/Pyrophosphate_aa/1'>Gly102, Arg103, Gly104, His105, Ser106 and Thr174</scene>. | + | The two negative charges brought by the pyrophosphate of the FAD cofactor is compensated by the nitrogen atoms of a few amino acids such as <scene name='68/686754/Pyrophosphate_aa/1'>Gly102, Arg103, Gly104, His105, Ser106 and Thr174</scene>.<ref name="Malitoa">PMID: 15321719 </ref> |
===='''Substrate binding domain'''==== | ===='''Substrate binding domain'''==== | ||
Revision as of 15:31, 9 January 2015
2qkn
Crystal structure of Maize cytokinin oxidase/dehydrogenase complexed with phenylurea inhibitor CPPU
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Frebortova J, Novak O, Frebort I, Jorda R. Degradation of cytokinins by maize cytokinin dehydrogenase is mediated by free radicals generated by enzymatic oxidation of natural benzoxazinones. Plant J. 2010 Feb 1;61(3):467-81. doi: 10.1111/j.1365-313X.2009.04071.x. Epub, 2009 Nov 14. PMID:19912568 doi:http://dx.doi.org/10.1111/j.1365-313X.2009.04071.x
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Kopecny D, Briozzo P, Popelkova H, Sebela M, Koncitikova R, Spichal L, Nisler J, Madzak C, Frebort I, Laloue M, Houba-Herin N. Phenyl- and benzylurea cytokinins as competitive inhibitors of cytokinin oxidase/dehydrogenase: a structural study. Biochimie. 2010 Aug;92(8):1052-62. Epub 2010 May 15. PMID:20478354 doi:10.1016/j.biochi.2010.05.006
- ↑ Kopecny D, Sebela M, Briozzo P, Spichal L, Houba-Herin N, Masek V, Joly N, Madzak C, Anzenbacher P, Laloue M. Mechanism-based inhibitors of cytokinin oxidase/dehydrogenase attack FAD cofactor. J Mol Biol. 2008 Jul 25;380(5):886-99. Epub 2008 May 24. PMID:18571199 doi:10.1016/j.jmb.2008.05.044
- ↑ 4.0 4.1 4.2 4.3 Kopecny D, Pethe C, Sebela M, Houba-Herin N, Madzak C, Majira A, Laloue M. High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie. 2005 Nov;87(11):1011-22. PMID:15927342 doi:http://dx.doi.org/10.1016/j.biochi.2005.04.006
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Malito E, Coda A, Bilyeu KD, Fraaije MW, Mattevi A. Structures of Michaelis and product complexes of plant cytokinin dehydrogenase: implications for flavoenzyme catalysis. J Mol Biol. 2004 Aug 27;341(5):1237-49. PMID:15321719 doi:http://dx.doi.org/10.1016/j.jmb.2004.06.083
- ↑ 6.0 6.1 Schmulling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res. 2003 Jun;116(3):241-52. Epub 2003 Apr 29. PMID:12721786 doi:http://dx.doi.org/10.1007/s10265-003-0096-4