We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 951
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
=====Light emission===== | =====Light emission===== | ||
Luciferase can react to emit light. In this reaction, luciferase firstly synthetized luciferin-AMP from luciferin and ATP, using a Mg2+ ion to offset the negative charges of the phosphate groups. Then, luciferase turns the luciferin-AMP into oxyluciferin in an excited state thanks to a dioxygen. This step releases AMP and CO2. The excited oxyluciferin relaxes and looses a photon, so light is emitted. | Luciferase can react to emit light. In this reaction, luciferase firstly synthetized luciferin-AMP from luciferin and ATP, using a Mg2+ ion to offset the negative charges of the phosphate groups. Then, luciferase turns the luciferin-AMP into oxyluciferin in an excited state thanks to a dioxygen. This step releases AMP and CO2. The excited oxyluciferin relaxes and looses a photon, so light is emitted. | ||
| - | The wavelength of the light can vary with the pH, which can be explained by the luciferase structure: at the physiological pH, the emitted light is green and at a lower pH, the color is red.<ref name = | + | The wavelength of the light can vary with the pH, which can be explained by the luciferase structure: at the physiological pH, the emitted light is green and at a lower pH, the color is red.<ref name ="first">PMID:8805533</ref> |
=====Fatty-acyl-CoA synthesis===== | =====Fatty-acyl-CoA synthesis===== | ||
This reaction uses similar reaction but instead of taking luciferin as ligand, it takes fatty acids. Using ATP-Mg2+, it firstly forms fatty-acyl-AMP. And then, CoA-SH attacks the carboxylic group of fatty-acyl-AMP and so forms fatty-acyl-CoA. | This reaction uses similar reaction but instead of taking luciferin as ligand, it takes fatty acids. Using ATP-Mg2+, it firstly forms fatty-acyl-AMP. And then, CoA-SH attacks the carboxylic group of fatty-acyl-AMP and so forms fatty-acyl-CoA. | ||
| Line 16: | Line 16: | ||
== Global Structure == | == Global Structure == | ||
| - | Luciferase is a 62kDa protein and contains 550 amino acids. This enzyme can be divide into two domains. On the one hand, the major portion, corresponding to the <scene name='60/604470/N_terminal_domain/1'>N-terminal</scene>. On the other hand, the small portion, corresponding to the <scene name='60/604470/C_terminal_domain/1'>C-terminal domain</scene>.Those two domains are separated by a large cleft.<ref name = | + | Luciferase is a 62kDa protein and contains 550 amino acids. This enzyme can be divide into two domains. On the one hand, the major portion, corresponding to the <scene name='60/604470/N_terminal_domain/1'>N-terminal</scene>. On the other hand, the small portion, corresponding to the <scene name='60/604470/C_terminal_domain/1'>C-terminal domain</scene>.Those two domains are separated by a large cleft.<ref name ="first">PMID:8805533</ref> |
====The C-terminal domain==== | ====The C-terminal domain==== | ||
| - | The sequence of amino acids of C-terminal domain is composed of contiguous residues and forms a type of lid upon the N-terminal domain. It contains two β sheets: the <scene name='60/604470/Beta_sheet/2'>first β sheet </scene> is composed of two short antiparallel strands and the <scene name='60/604470/Beta_sheet_2/1'> second β sheet</scene> is composed of 3 antiparallel strands, mixed with <scene name='60/604470/3_alpha_helices/1'>three α helices</scene>. Those helices are put toward the outside. It is α + β structure.<ref name = | + | The sequence of amino acids of C-terminal domain is composed of contiguous residues and forms a type of lid upon the N-terminal domain. It contains two β sheets: the <scene name='60/604470/Beta_sheet/2'>first β sheet </scene> is composed of two short antiparallel strands and the <scene name='60/604470/Beta_sheet_2/1'> second β sheet</scene> is composed of 3 antiparallel strands, mixed with <scene name='60/604470/3_alpha_helices/1'>three α helices</scene>. Those helices are put toward the outside. It is α + β structure.<ref name ="first">PMID:8805533</ref> |
====The N-terminal domain==== | ====The N-terminal domain==== | ||
| Line 34: | Line 34: | ||
The active site is not strictly highlighted according to the actual state of studies but some residues and motifs strongly modified have been determined and this conformation enables to find the active site. Many of these conserved residues are located on the core of the β-barrel, on the small C-terminal domain and in the surface of the N-terminal domain, which forms a <scene name='60/604470/Depression_c-ter_and_n-ter/1'>depression</scene>. However, this depression is too large to enable interactions between residues and substrates, so it is thought that a conformational change occurs and sandwiches the substrates, forming the active site. This conformational change provides a suitable environment for light production because of exclusion of water molecules from the active site, favouring intramolecular reactions. Residues also follow a <scene name='60/604470/Cleft/1'>cleft</scene> caused by of the <scene name='60/604470/Beta_sheet_b/3'>Beta-sheet B</scene> against the <scene name='60/604470/Beta_barrel/3'>Beta-barrel</scene>.<ref name =''first''>PMID:8805533</ref> | The active site is not strictly highlighted according to the actual state of studies but some residues and motifs strongly modified have been determined and this conformation enables to find the active site. Many of these conserved residues are located on the core of the β-barrel, on the small C-terminal domain and in the surface of the N-terminal domain, which forms a <scene name='60/604470/Depression_c-ter_and_n-ter/1'>depression</scene>. However, this depression is too large to enable interactions between residues and substrates, so it is thought that a conformational change occurs and sandwiches the substrates, forming the active site. This conformational change provides a suitable environment for light production because of exclusion of water molecules from the active site, favouring intramolecular reactions. Residues also follow a <scene name='60/604470/Cleft/1'>cleft</scene> caused by of the <scene name='60/604470/Beta_sheet_b/3'>Beta-sheet B</scene> against the <scene name='60/604470/Beta_barrel/3'>Beta-barrel</scene>.<ref name =''first''>PMID:8805533</ref> | ||
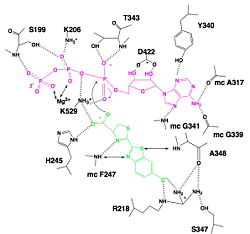
| - | [[Image:Luciferin_bounding_to_luciferase.jpg|250px|right|thumb|Hydrogen bonding between Luciferase and substrates luciferin (green), ATP (violet) and Mg2+,<ref name= | + | [[Image:Luciferin_bounding_to_luciferase.jpg|250px|right|thumb|Hydrogen bonding between Luciferase and substrates luciferin (green), ATP (violet) and Mg2+,<ref name="second">[http://www.photobiology.info/ Photobiology]</ref>]] |
=====Interaction with ATP===== | =====Interaction with ATP===== | ||
| - | We find a signal motif in luciferase which is <scene name='60/604470/Atp_binding_signal_motif/1'>[STG]-[STG]-G-[ST]-[ST]-[TSE]-[GS]-x-[PALIVM]-K</scene>. Here some residues like lysine are always conserved. This pattern enables ATP binding thanks to hydrogen bonds between residues and phosphates of ATP. There is another pattern : <scene name='60/604470/Adenosine_ring_binding/1'>[YFW]-[GASW]-x-[TSA]-E</scene> which takes a particular conformation because of hydrogen bonds between residues and maintain the adenosin ring of ATP.<ref name = | + | We find a signal motif in luciferase which is <scene name='60/604470/Atp_binding_signal_motif/1'>[STG]-[STG]-G-[ST]-[ST]-[TSE]-[GS]-x-[PALIVM]-K</scene>. Here some residues like lysine are always conserved. This pattern enables ATP binding thanks to hydrogen bonds between residues and phosphates of ATP. There is another pattern : <scene name='60/604470/Adenosine_ring_binding/1'>[YFW]-[GASW]-x-[TSA]-E</scene> which takes a particular conformation because of hydrogen bonds between residues and maintain the adenosin ring of ATP.<ref name ="first">PMID:8805533</ref>, <ref name="second">[http://www.photobiology.info/ Photobiology]</ref> |
=====Interaction with luciferin===== | =====Interaction with luciferin===== | ||
| - | Luciferase holds the luciferin with the specific residues <scene name='60/604470/Residues_helding_luciferin/1'>arginin 218, phenylalanin 247, serin 347 and adenin 348</scene>, still with hydrogen bounds. Those bindings make the carboxylate oxygen of luciferin points toward the α phosphate of ATP, so the oxygen is well-positionned to attack the α phosphate. This promotes the luciferin-AMP formation.<ref name = | + | Luciferase holds the luciferin with the specific residues <scene name='60/604470/Residues_helding_luciferin/1'>arginin 218, phenylalanin 247, serin 347 and adenin 348</scene>, still with hydrogen bounds. Those bindings make the carboxylate oxygen of luciferin points toward the α phosphate of ATP, so the oxygen is well-positionned to attack the α phosphate. This promotes the luciferin-AMP formation.<ref name ="first">PMID:8805533</ref>, <ref name="second">[http://www.photobiology.info/ Photobiology]</ref> |
=====Interaction with fatty acids===== | =====Interaction with fatty acids===== | ||
Fatty acids are highly similar to luciferin. Therefore, luciferase can use the luciferin binding site to bind fatty acids. That is why they can be used as substrates by luciferase and then, very high similar reaction as for luciferin occurs. | Fatty acids are highly similar to luciferin. Therefore, luciferase can use the luciferin binding site to bind fatty acids. That is why they can be used as substrates by luciferase and then, very high similar reaction as for luciferin occurs. | ||
Revision as of 18:43, 9 January 2015
| |||||||||||
References
- ↑ Welsh DK, Kay SA. Bioluminescence imaging in living organisms. Curr Opin Biotechnol. 2005 Feb;16(1):73-8. PMID:15722018 doi:http://dx.doi.org/10.1016/j.copbio.2004.12.006
- ↑ 2.0 2.1 2.2 2.3 2.4 Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ 4.0 4.1 Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ 5.0 5.1 5.2 Photobiology
- ↑ Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009 Jan;61(1):6-17. PMID:18949818 doi:10.1002/iub.134
- ↑ Hosseinkhani S. Molecular enigma of multicolor bioluminescence of firefly luciferase. Cell Mol Life Sci. 2011 Apr;68(7):1167-82. doi: 10.1007/s00018-010-0607-0. Epub, 2010 Dec 28. PMID:21188462 doi:http://dx.doi.org/10.1007/s00018-010-0607-0
- ↑ Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell Mol Life Sci. 2010 Feb;67(3):387-404. Epub 2009 Oct 27. PMID:19859663 doi:10.1007/s00018-009-0170-8