You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Context
(or IDE), is a human metallopeptidase. It plays an important role in the cellular degradation of many molecules. Its activity was first related only to insulin, but now, we know that IDE interacts with insulin, glugacon and bradykinin.
The surprising thing is that IDE can be in complex with bradykinin. In fact, bradykinin is a small peptide (9 amino acids), and generally, IDE has substrates longer than 20 amino acids.
Normally, IDE can not bind small molecules. That's why this complex is interessant to study. Bradykinin is a small ligand and can also interacts with the catalytic chamber of IDE.
The IDE-bradykinin complex has a complexity in the substrate binding and recognition mechanism of IDE.
Previous studies (???) reveal that IDE uses an exosite to interact with the N-terminal end of its substrate (???), away from the catalytic site. This particular catch allows multiple cleavages of substrates by IDE. It was also observed that the binding of bradykinin occurs at the exosite and not the catalytic site.
Insuline degrading enzyme (IDE)
(EC 3.4.24.56) is a human enzyme of the metallopeptidase family, not well-known yet. It is composed by more than 1000 residues and has a huge catalytic cavity. It is made of 2 parts linked by a loop, and it switches between an open and a close state. The size of its catalytic chamber allows the binding of peptides (70 amino acids long). IDE hydrolyzes a lot of substrates which have many differents biological activities. Its substrate can be insuline, glucagon, amyline or bradykinin.
This enzyme hydrolyses its substrates by cleaving them at different points. Substrates have similar secondary structures.
Exosite: an essential element for the catalysis
This is an essential component for the substrate hydrolysis. It plays a role in the docking of substrates on IDE. Actually, when substrates arrive, they first bind to the exosite and then, they are translocated to the catalytic site. They have to change their conformation in order to access to the catalytic site.
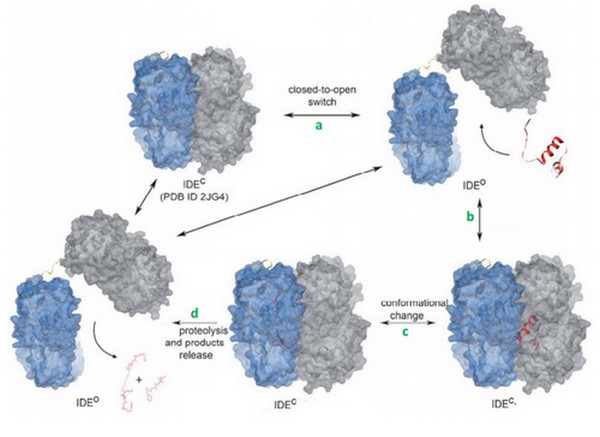
Catalytic mechanism
IDE can switch between its open and its close state. In the close state, IDE can not bind a substrate but it can hydrolyze it if it was already binding. In the other hand, in the open state, IDE can bind a substrate but it is less active because the catalytic site is not completly close.
The catalytic mechanism occurs in 4 steps:
1. IDE changes its state from close to open. This permitts the binding of the substrate in the catalytic chamber.
2. The IDE-substrate complex changes into close conformation.
3. The substrate can now bind to the exosite with his N-term tail and then change his conformation in order to be acceptable for the catalytic site. IDE can now cleave the substrate.
4. IDE changes into open conformation and release products.
Bradykinin
Bradykinin biological roles
is a short lived vasoactive nonapeptide mediator of the family of kinins. It's secreted in response to an inflammatory envent and serves as a mediator of pain, inflammation and vasodilatation.
Bradykinin synthesis
it is secreted by the enzymatic action of kallikreins on kininogen precursors (?). Kallidin is a produt of the enzymatic action of kallikrein to kininogens. Then Kallidin is transformed into bradykinin
Today, we know that kinins can be degraded by IDE.
We supposed that IDE may bind two molecules of bradykinin at the same time
Recognition and interactions between bradykinin and IDE
IDE can bind many substrates to its exosite and must recognize them. In order to accomplish this task, IDE uses many tools.
First, the lenght of the substrate peptide is essential. The peptide must be long enough to touch both the exosite and the catalytic site. Second, the peptide must be short enough in order to enter the catalytic chamber. In the case of the bradykinin, the length is too small to touch the catalytic site when it binds the exosite.
Tang et al have shown that bradykinin is too small to bind both at the exosite and the catalytic side. They prooved that IDE binds 2 bradykinin at the same time : the first one interacts with the exosite and the second one touches the catalytic site. The bradykinin in the catalytic site is the one which is going to be cleaved by the enzyme.
This can be a discriminate factor, because IDE binds 2 bradykinin at the same time.
IDE catalytic site has a high affinity for hydrophobic and basic. Bradykinin is essentially composed by proline and arginine, which are basic amino acids. So, bradykinin structure may explain this strange interaction.
Crystal structure revealed that and of IDE are involved in interactions with bradykinin.
N-ter 3 residues of bradykinin (Arg1, Pro2, Pro3) is also found to interact with the exosite.
Hypothetical role of bradykinin on IDE
Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state.
We can also suggests that IDE binds thanks to their small lenght.
Binding of bradykinin or other short peptides to the exosite could play a regulatory role in substrate binding and cleavage by IDE.
Bradykinin is supposed to reduce the size of the ctalytic chamber of IDE, and this may enhance the substrate binding and cleavage by reducing the entropy of short peptides in the chamber. Bradykinin is shown to be an activator of IDE.
In the other hand, it could reduce the cleavage of other substrates by interfering with their binding.
The degradation of kinins and specially bradykinin is not well understand yet. The cleavage site of bradykinin by IDE, the detailed kinetic analysis and the structural basis for recognition have to be more studied.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

