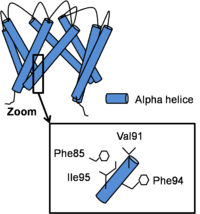Introduction
Ion channels are transmembrane proteins which allow ions to pass through biological membranes.
Some of these channels are very selective, others have a low level of selectivity. The NaK channel is a
non-selective one : It conduits cations more than anions but it let pass several cations : Na+, K+, Rb+, and Ca2+ [1]
Understanding how these channels work is important because in the organism a lot of messages are transmitted through electric currents (which are ionic currents across the membrane) : nerves impulse, photoreceptors, etc. Thus, these not very selective NaK channels are very interesting for the inhibition of intercellular messages for instance.
Structure
General Description
The NaK channel is like an intracellular gate.
The NaK channel have the same general architecture as the K+ channels. In fact, the NaK channel has 4 subunits which are symmetric with respect to the central axis of the pore. Each subunit is composed of 3 alpha-helices. One of them is a short pore helix which is oblique to the channel axis. The others are the outer and the inner helices and they extend across the lipid membrane. are around the structure so the helices can cross the membrane
Structure of the open or closed complex
In response to a external stimuli, the structure of the NaK channel is different. In fact, after some inter- and intra-subunit rearrangements, the NaK channel can be open or closed.

The structure of the channel in his closed conformation Closed Conformation
In the closed conformation, inner helices are near and straight. There is a subsequent bundle crossing formed by interactions between C-terminal residues. In the region just above the bundle crossing, from each inner helix forms contacts with a hydrophobic patch on the opposite face of Phe92 from the neighboring inner helix formed by Val91, Phe94, Ile95 and Leu98. [2]
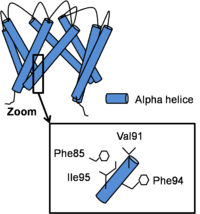
The structure of the channel in his open conformation Open Conformation
Channel opening is a conserved mechanism.The inner helix twist and bend thanks to a which is considered as the gating hinge. After this bending, the inner helices twist of 45° around their helical helix and the outer helix tilt tangentially in the same direction by 11° without any twisting motion. As all of helix twist or move inside of a subunit, intra-subunit interactions between inner and outer helix don’t differ a lot. On the contrary, inter-subunit interactions between neighboring inner helix change. In fact, Phe92 swings away and points its side chain towards the central ion conduction pathway due to inner helix bending and the hydrophobic patch slides along the neighboring inner helix by two helical turns and forms new Van der Waals contacts with . This resulted in a disruption of the bundle crossing and so intra- and inter- subunits interactions in the open state become less important than in the close state. [3]
Active Site & Ions Passage
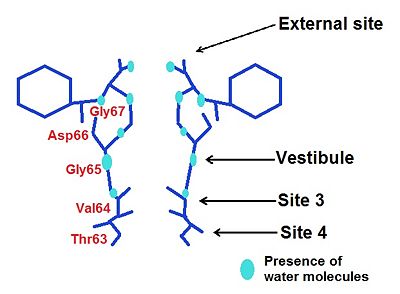
There are
4 ions binding sites in the NaK channel
[4]. This diversity allows by different mechanisms to conduit . They have similar chemical environments but they have
different ion selectivity. Two of them (sites S3 and S4) are conserved, that is to say they are the same than in the high selective K+ channel while S1 and S2 become a vestibular structure where K+ and Na+ ions can diffuse
[5] .
The different ions binding sites We will see for every binding site how his structure allows the passage of one or several ions.
External Site
We may notice the presence of a glycine () which brings four carbonyl oxygen atoms, more inward oriented, able to bind with water molecules. This create an environement which can chelate K+ and Rb+ ions, but avoid the binding of Na+.
Moreover, thanks to a space intercation between Asp 66 and Gly67, the external site has a higher affinity for divalent cations such as Ca2+ and Ba2+ rather than monovalent such as K+ and Rb+ [6].
The nature of the ligands is carbonyl-water.
Vestibule
In the case of the vestibule, there are too four carbonyl oxygen atom which brings by a valine (Val64). For instance, Na+ is neared to the ligand by this way: distance Na+-ligand=2,9 Ä. Moreover, ions are partially hydrated by four water molecules (they are along with the carboxyl oxygene atoms) : distance ions-H2O=4 Ä. The presence of water allows a greater flexibility in the ion binding so the vestibule may adapt to monovalent cations such as Na+, K+ and Rb+. However, this structure has a greater selectivity for K+ than Na+ : water molecules help to create a selectivity filter thanks to ligand geometry: octahedral arrangement which is impossible with Na+ because of a smaller radius and a hydratation by 5-6 molecules of water [7].
The nature of the ligands is carbonyl-water.
Site 3
He is the most non selective ion binding site which let pass mono and divalent cations, so a contamination can occur : presence of unkonwn species of ion at this site.
Moreover we may underscore a higher affinity for K+ than Na+ because of several reason :
First, we can find 4 backbone carbonyl oxygen from which participate in K+ and Rb+ ions chelation because of the formation of an octahedral ligand: an octahedral arrangement oxygen ligands in the channel pore is more favorable for K+ than Na+.
The lack of selectivity is due to the fact that the NaK channel have an almost identical structure when it is in complex with Na+, K+ or Rb+ : there is no big rearrangement in the structure of the protein depending on the bound ion. So the structure is stable with any ions, so it is non selective. Moreover, it could have a heavy atom contamination but it happens in a smaller extent with K+ than with Na+.
The amino-acids of the site 3 participate a lot in the transfert of Na+. In this case, Na+ binds because of an H-bonding interactions between and the backbone amide of which stabilize the structure. Furthermore, and form a ion binding cage where Na+ ions tend bind at upper or lower ends (not in the center).
The nature of the ligands are carbonyl-carbonyl.
Site 4
We find again a ion binding cage made by carbonyl oxygen atoms from . Na+ ions have almost a planar conformation with respect to its ligands : distance of 2,4 Ä with the four hydroxyl oxygen atoms. There is also a coordination with water molecule in the central cavity : distance of 2,7 Ä between H2O and Na+.
The nature of the ligands is carbonyl-hydroxyl.
It seems that Na+ binding positions are site 3 and 4 where the amino-acids form a cage in which the ions are chelated in plane with their ligands.
We observe that NaK filter is able to bind both Na+ and K+ thanks to existing environment rather than structural rearrangements.
The Filter Selectivity
The filter is defined by a that’s why the channel is selective for some cations like K+ or Na+. The selectivity filter has the same conformation in low K+/high Na+ or high K+/low Na+ concentrations. So the concentration does not impact the conformation of the filter but it can adopt 2 different structures : a conductive state and a non conductive state. In fact, some hydrogen bonds are important for the stability of the NaK selectivity filter and the balance between the 2 structures. For example, an hydrogen bond between residues and stabilize the non conductive state whereas an hydrogen bond between and stabilize the conductive state. The change between the 2 structures are very fast.
The conductive state is characterized by a low energy barrier. In contrast, a non conductive state is characterized by high energy barrier.
References
- ↑ PMC3183810
- ↑ Alam A, Jiang Y. High-resolution structure of the open NaK channel. Nat Struct Mol Biol. 2009 Jan;16(1):30-4. Epub 2008 Dec 21. PMID:19098917 doi:nsmb.1531
- ↑ Alam A, Jiang Y. High-resolution structure of the open NaK channel. Nat Struct Mol Biol. 2009 Jan;16(1):30-4. Epub 2008 Dec 21. PMID:19098917 doi:nsmb.1531
- ↑ Shi N, Ye S, Alam A, Chen L, Jiang Y. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006 Mar 23;440(7083):570-4. Epub 2006 Feb 8. PMID:16467789 doi:10.1038/nature04508
- ↑ Alam A, Jiang Y. Structural analysis of ion selectivity in the NaK channel. Nat Struct Mol Biol. 2009 Jan;16(1):35-41. Epub 2008 Dec 21. PMID:19098915 doi:nsmb.1537
- ↑ Alam A, Shi N, Jiang Y. Structural insight into Ca2+ specificity in tetrameric cation channels. Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15334-9. Epub 2007 Sep 18. PMID:17878296
- ↑ Varma S, Rempe SB. Coordination numbers of alkali metal ions in aqueous solutions. Biophys Chem. 2006 Dec 1;124(3):192-9. Epub 2006 Jul 27. PMID:16875774 doi:http://dx.doi.org/10.1016/j.bpc.2006.07.002
Contributors
Camille Noblet & Lola Welsch