We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 972
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
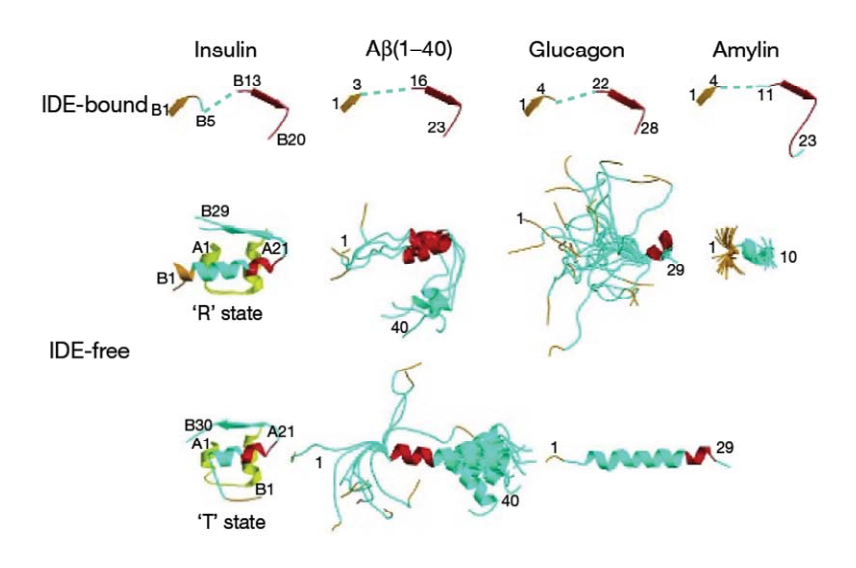
First, the lenght of the substrate peptide is essential. The peptide must be long enough to touch both the exosite and the catalytic site. Second, the peptide must be short enough in order to enter the catalytic chamber. In the case of the bradykinin, the length is too small to touch the catalytic site when it binds the exosite. | First, the lenght of the substrate peptide is essential. The peptide must be long enough to touch both the exosite and the catalytic site. Second, the peptide must be short enough in order to enter the catalytic chamber. In the case of the bradykinin, the length is too small to touch the catalytic site when it binds the exosite. | ||
Song et al<ref>doi:10.1074/jbc.M308983200</ref> have shown that bradykinin is too small to bind both at the exosite and the catalytic side. They prooved that IDE binds 2 bradykinin at the same time : the first one interacts with the exosite and the second one touches the catalytic site. The bradykinin in the catalytic site is the one which is going to be cleaved by the enzyme. | Song et al<ref>doi:10.1074/jbc.M308983200</ref> have shown that bradykinin is too small to bind both at the exosite and the catalytic side. They prooved that IDE binds 2 bradykinin at the same time : the first one interacts with the exosite and the second one touches the catalytic site. The bradykinin in the catalytic site is the one which is going to be cleaved by the enzyme. | ||
| - | This can be a discriminate factor, because IDE binds <scene name='60/604491/Bradykinin/1'>2 bradykinins</scene> at the same time. | ||
IDE catalytic site has a high affinity for hydrophobic and basic. Bradykinin is essentially composed by proline and arginine, which are basic amino acids. So, bradykinin structure may explain this strange interaction. | IDE catalytic site has a high affinity for hydrophobic and basic. Bradykinin is essentially composed by proline and arginine, which are basic amino acids. So, bradykinin structure may explain this strange interaction. | ||
| Line 34: | Line 33: | ||
N-ter 3 residues of bradykinin (<scene name='60/604491/Bradyn-ter/1'>Arg1, Pro2, Pro3</scene>) is also found to interact with the exosite. | N-ter 3 residues of bradykinin (<scene name='60/604491/Bradyn-ter/1'>Arg1, Pro2, Pro3</scene>) is also found to interact with the exosite. | ||
| - | + | The degradation of kinins and specially bradykinin is not well understand yet. The cleavage site of bradykinin by IDE, the detailed kinetic analysis and the structural basis for recognition have to be more studied. | |
==Hypothetical role of bradykinin on IDE== | ==Hypothetical role of bradykinin on IDE== | ||
| - | + | Bradykinin is well known as an activator of IDE (34). Consequently, the binding of one bradykinin at the exosite must have an influence on the binding of other substrates. Actually, small peptides like bradykinin can reduce the catalytic chamber by the binding to the exosite. This leads to an increase of the binding of other substrates and the subsequent cleavage by IDE. Alternatively, bradykinin binding could reduce the cleavage by interfering with substrate binding<ref></ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
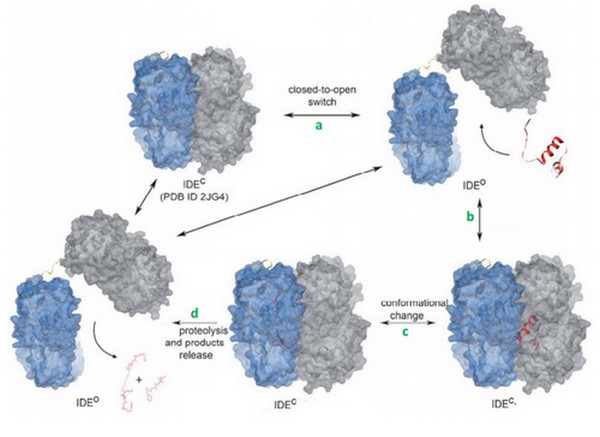
| - | + | Today, we can supposed that binding of bradykinin at the exosite stimulated the conformationnal change of IDE, from its open to its close state. Binding of bradykinin or other short peptides to the exosite could play a regulatory role in substrate binding and cleavage by IDE<ref>doi: 10.1021/bi801192h</ref>. | |
| + | |||
| + | Im et al<ref>doi: 10.1074/jbc.M701590200</ref> suggested that ATP increases the activtity of IDE with its small substrates like braydkinin. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 22:32, 9 January 2015
| This Sandbox is Reserved from 15/11/2014, through 15/05/2015 for use in the course "Biomolecule" taught by Bruno Kieffer at the Strasbourg University. This reservation includes Sandbox Reserved 951 through Sandbox Reserved 975. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007 Aug 31;282(35):25453-63. Epub 2007 Jul 5. PMID:17613531 doi:10.1074/jbc.M701590200
- ↑ Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006 Oct 19;443(7113):870-4. Epub 2006 Oct 11. PMID:17051221 doi:10.1038/nature05143
- ↑ Song ES, Juliano MA, Juliano L, Hersh LB. Substrate activation of insulin-degrading enzyme (insulysin). A potential target for drug development. J Biol Chem. 2003 Dec 12;278(50):49789-94. Epub 2003 Oct 2. PMID:14527953 doi:http://dx.doi.org/10.1074/jbc.M308983200
- ↑ Malito E, Ralat LA, Manolopoulou M, Tsay JL, Wadlington NL, Tang WJ. Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme. Biochemistry. 2008 Nov 6. PMID:18986166 doi:10.1021/bi801192h
- ↑ Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007 Aug 31;282(35):25453-63. Epub 2007 Jul 5. PMID:17613531 doi:10.1074/jbc.M701590200