Mycobacterium tuberculosis ArfA Rv0899
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
==Introduction== | ==Introduction== | ||
| - | The Rv0899 ArfA protein [[http://www.uniprot.org/uniprot/P9WIU5 ARFA_MYCTU]] from [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis] | + | The Rv0899 ArfA protein [[http://www.uniprot.org/uniprot/P9WIU5 ARFA_MYCTU]] from [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis] H37Rv belongs to the OmpA (outer membrane protein A) family of outer membrane proteins and has been proposed to act as an outer membrane [[porin]] with ion channel activity and contributes to the bacterium's adaptation to the acidic environment in phagosome [http://en.wikipedia.org/wiki/Phagosome] during infection <ref>PMID: 21117233 </ref>. Rv0899 protein encoded by an ammonia release facilator operon that is necessary for rapid ammonia secretion, pH neutralization and adaptation to acidic environments in vitro. Two ''Mycobacterium tuberculosis'' H37Rv genes (Rv0900 [[http://www.uniprot.org/uniprot/P9WJG7 ARFB_MYCTU]] and Rv0901 [[http://www.uniprot.org/uniprot/P9WJG5 ARFC_MYCTU]] ) adjacent to Rv0899 also encode putative membrane proteins, and are found exclusively in association with Rv0899 in the same pathogenic mycobacteria, suggesting that the three may constitute an operon dedicated to a common function. |
| - | Asparagine is the primary ammonia source for Mycobacterium tuberculosis H37Rv at acidic pH <ref>PMID: 21410778 </ref>. | + | Asparagine is the primary ammonia source for ''Mycobacterium tuberculosis'' H37Rv at acidic pH <ref>PMID: 21410778 </ref>. |
The deletion of this gene impairs the uptake of some water-soluble substances, such as serine, glucose, and glycerol <ref>PMID: 12366842 </ref>. | The deletion of this gene impairs the uptake of some water-soluble substances, such as serine, glucose, and glycerol <ref>PMID: 12366842 </ref>. | ||
| Line 20: | Line 20: | ||
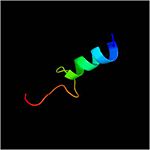
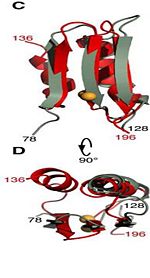
Residues <scene name='61/612805/N-c_rainbow/1'>73-326</scene> form a mixed alpha/beta-globular structure, encompassing two independently folded modules corresponding to the B and C domains connected by a flexible linker. | Residues <scene name='61/612805/N-c_rainbow/1'>73-326</scene> form a mixed alpha/beta-globular structure, encompassing two independently folded modules corresponding to the B and C domains connected by a flexible linker. | ||
| - | The central B domain | + | The central B domain <scene name='61/612805/Secondary_structure_b/1'>residues 73-200</scene>> folds with three parallel/antiparallel {{Template:ColorKey_helixes}} packed against six parallel/antiparallel {{Template:ColorKey_strands}} that form a flat beta-sheet. The B domain has homology with conserved putative <scene name='61/612805/Conserved_g95_and_g164_in_bon/2'>lipid-binding BON</scene> (bacterial OsmY and nodulation) superfamily domains and conserved Gly95 and Gly164 [http://www.ebi.ac.uk/interpro/entry/IPR014004]. <scene name='61/612805/Surface2/1'>The core</scene> is {{Template:ColorKey_Hydrophobic}}, while the exterior is {{Template:ColorKey_Polar}} and predominantly acidic. The two subdomains are symmetric about α2, and their backbone atoms can be aligned with a RMSD [http://http://en.wikipedia.org/wiki/Root-mean-square_deviation] of 1.8Å, by performing a 180° rotation of either one around an axis normal to the 6-stranded β-sheet. |
Revision as of 21:34, 23 January 2015
| |||||||||||