Mycobacterium tuberculosis ArfA Rv0899
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
2. Bacterium's adaptation to the acidic environment (stress response) of the phagosome during infection by: | 2. Bacterium's adaptation to the acidic environment (stress response) of the phagosome during infection by: | ||
a) deamidation of the amino acid pair <scene name='61/612805/Asn111_and_gly112/1'> Asn111-Gly112 </scene>, located at the end of α1 and preceding L3, a pH-dependent reaction whereby Asn is converted to Asp and ammonia is released. Asparagine residues preceding glycine, and situated in conformationally flexible regions of proteins, are frequently deamidated, with potentially significant consequences for protein regulation and function <ref>PMID: 20199110</ref>. [[Image:Asparaginase-reaction.jpg|250px]] | a) deamidation of the amino acid pair <scene name='61/612805/Asn111_and_gly112/1'> Asn111-Gly112 </scene>, located at the end of α1 and preceding L3, a pH-dependent reaction whereby Asn is converted to Asp and ammonia is released. Asparagine residues preceding glycine, and situated in conformationally flexible regions of proteins, are frequently deamidated, with potentially significant consequences for protein regulation and function <ref>PMID: 20199110</ref>. [[Image:Asparaginase-reaction.jpg|250px]] | ||
| - | |||
b) pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | b) pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | ||
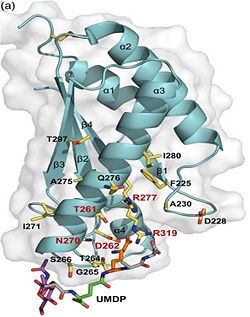
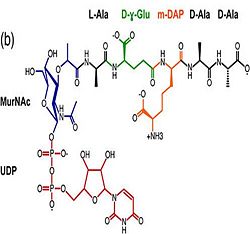
| - | 3. Contribution for structural strength to the bacterial cell wall ander acid or other stress conditionsIts functions by binding to peptidoglycan biosyntheesis intermediate uridine-5-'-diphosphate-MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala [http://en.wikipedia.org/wiki/Peptidoglycan] binding site <scene name='61/612805/Peptidoglycan_binding_site/1'>(R277, R319, T261, D262, N270)</scene>. These residues are strictly conserved in the OmpA -like family <ref>PMID: 22206986 </ref>. | + | 3. Contribution for structural strength to the bacterial cell wall ander acid or other stress conditionsIts functions by binding to peptidoglycan biosyntheesis intermediate uridine-5-'-diphosphate-MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala (UMDP) [http://en.wikipedia.org/wiki/Peptidoglycan] binding site <scene name='61/612805/Peptidoglycan_binding_site/1'>(R277, R319, T261, D262, N270)</scene>. These residues are strictly conserved in the OmpA -like family The side chain of m-DAP is stabilized by charge-charge interactions between its electronegative carbonyl group with R277 and R319 guanidinium groups and with the N270 carboxamide and between its electropositive amino group with the D262 carboxyl and the T261 hydroxyl. UMDP could interact through contacts of its γ-Glu3 and Ala2 backbone amides with the side-chain hydroxyl of S266, of its MurNAc O3 with the amide proton of E267, and of its MurNAc NH2 with the E267 carboxyl <ref>PMID: 22206986 </ref>. |
[[Image:123456.jpg|250px]] [[Image:MurNac11.jpg|250px]] | [[Image:123456.jpg|250px]] [[Image:MurNac11.jpg|250px]] | ||
Revision as of 10:36, 24 January 2015
| |||||||||||