Vitis vinifera Flavonoid 3-O-Glucosyltransferase (Vv3GT)
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
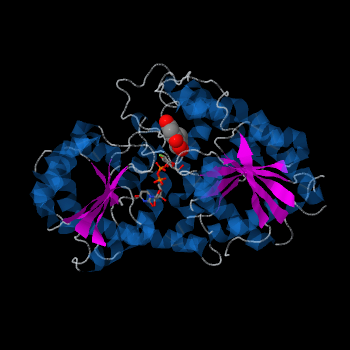
Despite low primary sequence similarity, the secondary and tertiary structures of GTs are highly conserved. <ref>PMID:16482224</ref> | Despite low primary sequence similarity, the secondary and tertiary structures of GTs are highly conserved. <ref>PMID:16482224</ref> | ||
| + | |||
| + | === GT-B === | ||
| + | |||
| + | === PSPG === | ||
| + | |||
| + | |||
| + | |||
| + | |||
Vv3GT is a GT-B enzyme. GT-B enzymes consists of two β/α/β Rossmann-like domains. The two domains are associated and face each other with the active-site lying between them. These domains are associated with the donor and acceptor substrate binding sites. | Vv3GT is a GT-B enzyme. GT-B enzymes consists of two β/α/β Rossmann-like domains. The two domains are associated and face each other with the active-site lying between them. These domains are associated with the donor and acceptor substrate binding sites. | ||
| Line 27: | Line 35: | ||
Three conserved motifs involved in sugar binding are present in Vv3GT. The first, a <scene name='60/607848/2c1z_loop_n5_label/1'>loopN5</scene> motif (Thr 141; Ala 142) involved in sugar binding. The second, a <scene name='69/692252/2c1z_wns_label/1'>WNS</scene> (Trp 354; Asn 355; Ser 356) motif residues are involved in binding UDP phosphates. The third, <scene name='69/692252/2c1z_d_eq_label/1'>D/EQ</scene> motif residues also involved in sugar binding (Asp 374; Gln 375). The WNS and D/EQ motifs are part of the highly conserved PSPG region. | Three conserved motifs involved in sugar binding are present in Vv3GT. The first, a <scene name='60/607848/2c1z_loop_n5_label/1'>loopN5</scene> motif (Thr 141; Ala 142) involved in sugar binding. The second, a <scene name='69/692252/2c1z_wns_label/1'>WNS</scene> (Trp 354; Asn 355; Ser 356) motif residues are involved in binding UDP phosphates. The third, <scene name='69/692252/2c1z_d_eq_label/1'>D/EQ</scene> motif residues also involved in sugar binding (Asp 374; Gln 375). The WNS and D/EQ motifs are part of the highly conserved PSPG region. | ||
| - | + | == Quiz == | |
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 13:45, 24 January 2015
| |||||||||||
References
- ↑ Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006 Mar 22;25(6):1396-405. Epub 2006 Feb 16. PMID:16482224