Mycobacterium tuberculosis ArfA Rv0899
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
==Putative functions== | ==Putative functions== | ||
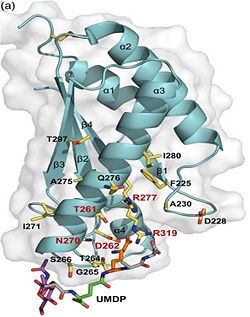
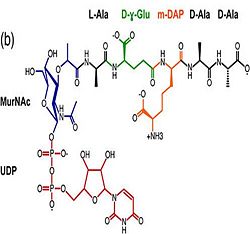
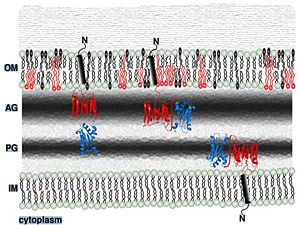
| - | 1. Contribution for structural strength to the bacterial cell wall ander acid or other stress conditionsIts functions by binding to peptidoglycan biosyntheesis intermediate uridine-5-'-diphosphate-MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala (UMDP) [http://en.wikipedia.org/wiki/Peptidoglycan] binding site. <scene name='61/612805/Peptidoglycan_binding_site/1'>(R277, R319, T261, D262, N270)</scene>. These residues are strictly conserved in the OmpA -like family The side chain of m-DAP is stabilized by charge-charge interactions between its electronegative carbonyl group with R277 and R319 guanidinium groups and with the N270 carboxamide and between its electropositive amino group with the D262 carboxyl and the T261 hydroxyl. UMDP could interact through contacts of its γ-Glu3 and Ala2 backbone amides with the side-chain hydroxyl of S266, of its MurNAc O3 with the amide proton of E267, and of its MurNAc NH2 with the E267 carboxyl <ref>PMID: 22206986 </ref> . | + | 1. Contribution for structural strength to the bacterial cell wall ander acid or other stress conditionsIts functions by binding to peptidoglycan biosyntheesis intermediate uridine-5-'-diphosphate-MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala (UMDP) [http://en.wikipedia.org/wiki/Peptidoglycan] binding site. <scene name='61/612805/Peptidoglycan_binding_site/1'>(R277, R319, T261, D262, N270)</scene>. These residues are strictly conserved in the OmpA -like family The side chain of m-DAP is stabilized by charge-charge interactions between its electronegative carbonyl group with R277 and R319 guanidinium groups and with the N270 carboxamide and between its electropositive amino group with the D262 carboxyl and the T261 hydroxyl. UMDP could interact through contacts of its γ-Glu3 and Ala2 backbone amides with the side-chain hydroxyl of S266, of its MurNAc O3 with the amide proton of E267, and of its MurNAc NH2 with the E267 carboxyl <ref>PMID: 22206986 </ref>. |
| + | |||
| + | [[Image:123456.jpg|250px]] [[Image:MurNac11.jpg|250px]] | ||
| + | |||
2. Acquisition of Zn(2+) ions by {{Template:ColorKey Composition Ligand}} <scene name='61/612805/Binding-site_for_zn/1'>L82, D96, F97, H125, D127 and V129</scene> <ref>PMID: 22108166 </ref>. | 2. Acquisition of Zn(2+) ions by {{Template:ColorKey Composition Ligand}} <scene name='61/612805/Binding-site_for_zn/1'>L82, D96, F97, H125, D127 and V129</scene> <ref>PMID: 22108166 </ref>. | ||
| Line 36: | Line 39: | ||
b) pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | b) pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | ||
| - | |||
| - | |||
| - | [[Image:123456.jpg|250px]] [[Image:MurNac11.jpg|250px]] | ||
4. Nitrogen fixation and / or nitrogen metabolism <ref>PMID: 12878000 </ref>. | 4. Nitrogen fixation and / or nitrogen metabolism <ref>PMID: 12878000 </ref>. | ||
Revision as of 16:06, 24 January 2015
| |||||||||||