This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Mycobacterium tuberculosis ArfA Rv0899
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
1. Acquisition of Zn(2+) ions by {{Template:ColorKey Composition Ligand}} <scene name='61/612805/Binding-site_for_zn/1'>binding-site L82, D96, F97, H125, D127 and V129</scene> for DNA replication and transcription <ref>PMID: 22108166 </ref>. | 1. Acquisition of Zn(2+) ions by {{Template:ColorKey Composition Ligand}} <scene name='61/612805/Binding-site_for_zn/1'>binding-site L82, D96, F97, H125, D127 and V129</scene> for DNA replication and transcription <ref>PMID: 22108166 </ref>. | ||
| - | |||
2. Rapid ammonia secretion to external environment: | 2. Rapid ammonia secretion to external environment: | ||
deamidation of the amino acid pair <scene name='61/612805/Asn111_and_gly112/1'> Asn111-Gly112 </scene>, located at the end of α1 and preceding L3, a pH-dependent reaction whereby Asn is converted to Asp and ammonia is released. Asparagine residues preceding glycine, and situated in conformationally flexible regions of proteins, are frequently deamidated, with potentially significant consequences for protein regulation and function <ref>PMID: 20199110</ref>. [[Image:Asparaginase-reaction.jpg|250px]] | deamidation of the amino acid pair <scene name='61/612805/Asn111_and_gly112/1'> Asn111-Gly112 </scene>, located at the end of α1 and preceding L3, a pH-dependent reaction whereby Asn is converted to Asp and ammonia is released. Asparagine residues preceding glycine, and situated in conformationally flexible regions of proteins, are frequently deamidated, with potentially significant consequences for protein regulation and function <ref>PMID: 20199110</ref>. [[Image:Asparaginase-reaction.jpg|250px]] | ||
| - | |||
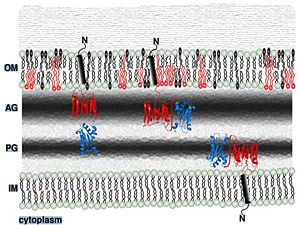
3. Stabilization of the outer membrane: | 3. Stabilization of the outer membrane: | ||
1. pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | 1. pH-dependent conformational dynamics of hydrophobic cluster of L232, F225, L240, A244, V281, L285 <scene name='61/612805/D236_before_mutation/1'>in neutral pH (D236) </scene> that folds to a more ordered structure like a flap at <scene name='61/612805/D236a_after_mut/3'>acidic pH (D236A) </scene>. | ||
| + | |||
2. binding to peptidoglycan and phospholipid layer. | 2. binding to peptidoglycan and phospholipid layer. | ||
Revision as of 15:48, 25 January 2015
| |||||||||||