Vitis vinifera Flavonoid 3-O-Glucosyltransferase (Vv3GT)
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
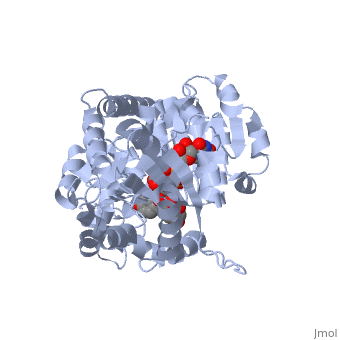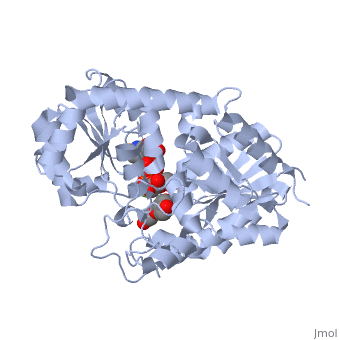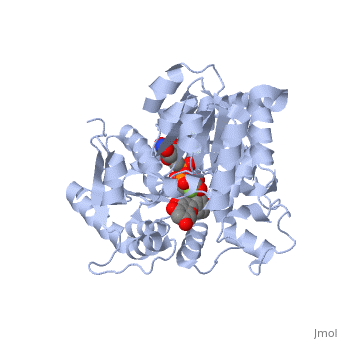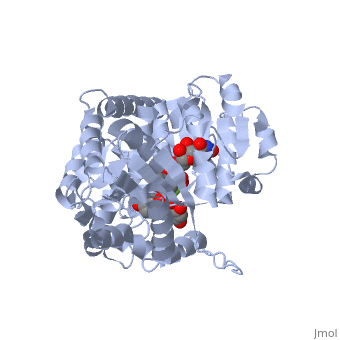
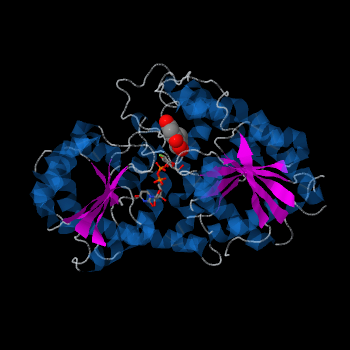
The <scene name='69/692252/2c1z_rainbow/1'>N-terminal and C-terminal domain</scene>, linked by a flexible loop and α-helix with a hinge region. The linker could be important for sugar donor binding. This linker region holds the UDP-sugar donor next to the C-terminal PSPG motif. The C-terminal domain is highly conserved and binds the UDP sugar donor (e. g. UDP-Glc). On the other hand, the N-terminal domain is not conserved it binds the substrate and provides catalytically active amino acids. | The <scene name='69/692252/2c1z_rainbow/1'>N-terminal and C-terminal domain</scene>, linked by a flexible loop and α-helix with a hinge region. The linker could be important for sugar donor binding. This linker region holds the UDP-sugar donor next to the C-terminal PSPG motif. The C-terminal domain is highly conserved and binds the UDP sugar donor (e. g. UDP-Glc). On the other hand, the N-terminal domain is not conserved it binds the substrate and provides catalytically active amino acids. | ||
| - | + | <table cellpadding="5" cellspacing="0"> | |
| + | <tr> | ||
| + | <td bgcolor="#0000ff"><font color="#ffffff" face="sans-serif"><b>N, 5'</b></font></td> | ||
| + | <td bgcolor="#0080ff"> </td> | ||
| + | <td bgcolor="#00c0ff"> </td> | ||
| + | <td bgcolor="#00e0b0"> </td> | ||
| + | <td bgcolor="#50e000"> </td> | ||
| + | <td bgcolor="#a0e000"> </td> | ||
| + | <td bgcolor="#e0e000"> </td> | ||
| + | <td bgcolor="#ffa000"> </td> | ||
| + | <td bgcolor="#ff0000"><font color="#ffffff" face="sans-serif"><b>C, 3'</b></font></td> | ||
| + | </tr></table> | ||
| + | |||
Revision as of 19:14, 25 January 2015
| |||||||||||
References
- ↑ Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011 Apr;66(1):182-93. doi: 10.1111/j.1365-313X.2011.04493.x. PMID:21443631 doi:http://dx.doi.org/10.1111/j.1365-313X.2011.04493.x
- ↑ Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, Sumner LW, Marin FR, Lewinsohn E, Fluhr R, Gressel J, Eyal Y. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 2004 Oct;40(1):88-100. PMID:15361143 doi:http://dx.doi.org/10.1111/j.1365-313X.2004.02193.x
- ↑ Osmani SA, Bak S, Moller BL. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry. 2009 Feb;70(3):325-47. doi: 10.1016/j.phytochem.2008.12.009. Epub, 2009 Feb 13. PMID:19217634 doi:http://dx.doi.org/10.1016/j.phytochem.2008.12.009
- ↑ Osmani SA, Bak S, Moller BL. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry. 2009 Feb;70(3):325-47. doi: 10.1016/j.phytochem.2008.12.009. Epub, 2009 Feb 13. PMID:19217634 doi:http://dx.doi.org/10.1016/j.phytochem.2008.12.009
- ↑ Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006 Mar 22;25(6):1396-405. Epub 2006 Feb 16. PMID:16482224