Binding site of AChR
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail Lymnaea stagnalis. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref>PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref>PMID:11683996</ref> | The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail Lymnaea stagnalis. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref>PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref>PMID:11683996</ref> | ||
| - | |||
| - | If we want to use the complex between α-bungarotoxin and the 13 amino acids to identify the binding site of the AChR, we should make sure that the little peptide has almost the same structure with the α-subunits of nAChR. From above we know that AChBP can be a model to study the structure of AChR. And the structure of AChBP has already been solved. So it easy to compare the little peptide with the AChBP. | ||
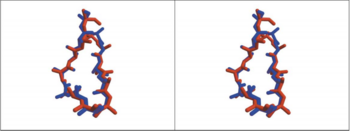
[[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. 1. Comparison, in Stere, of the 3D Structure of HAP(Red) and Loop 182-193 of AChBP(Blue)]] | [[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. 1. Comparison, in Stere, of the 3D Structure of HAP(Red) and Loop 182-193 of AChBP(Blue)]] | ||
| + | If we want to use the complex between α-bungarotoxin and the 13 amino acids to identify the binding site of the AChR, we should make sure that the little peptide has almost the same structure with the α-subunits of nAChR. From above we know that AChBP can be a model to study the structure of AChR. And the structure of AChBP has already been solved. So it easy to compare the little peptide with the AChBP. | ||
| + | |||
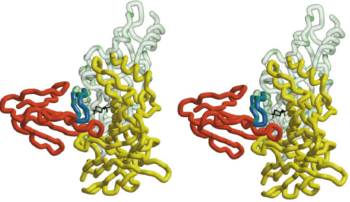
| + | [[Image:Combined BTX HAP and AchBP.png|thumb|350px|Fig. 2. A Stereo View of the Combined Model of α-BTX-HAP(Red) and AChBP subunits]] | ||
The overly of the first 12 residues of the 13-mer HAP on AChBP residues 182-193 shows that the HAP has almost the same conformation with the loop of AChBP(Fig 1), in the figure the red one is 13-mer little peptide and the blue one is a loop of AChBP. So it seems that the little peptide has almost the same structure with the region of AChR upon binding to α-BTX. | The overly of the first 12 residues of the 13-mer HAP on AChBP residues 182-193 shows that the HAP has almost the same conformation with the loop of AChBP(Fig 1), in the figure the red one is 13-mer little peptide and the blue one is a loop of AChBP. So it seems that the little peptide has almost the same structure with the region of AChR upon binding to α-BTX. | ||
| - | + | ||
The figure 2 shows that the <scene name='68/688431/Btx_complex_with_two_subunits/1'>superposition of the HAP on loop 182-193</scene> the α-BTX to fit exquisitely into the interface of two subunits of the pentameric AChBP. it shows the stereo view of the combined model of α-BTX-HAP(Red) and AChBP structure with subunit A in green and subunit B in yellow showing the insertion of loop 2 of the toxin into the interface of the to subunits.In order to identify more clearly that the little 13-mer peptide is actually have almost the same structure with the 182-193 loop with AChBP, we compare two structures: <scene name='68/688431/Btx_complex_with_two_subunits/5'>removing</scene> the HAP form the structure and <scene name='68/688431/Btx_complex_with_two_subunits/6'> | The figure 2 shows that the <scene name='68/688431/Btx_complex_with_two_subunits/1'>superposition of the HAP on loop 182-193</scene> the α-BTX to fit exquisitely into the interface of two subunits of the pentameric AChBP. it shows the stereo view of the combined model of α-BTX-HAP(Red) and AChBP structure with subunit A in green and subunit B in yellow showing the insertion of loop 2 of the toxin into the interface of the to subunits.In order to identify more clearly that the little 13-mer peptide is actually have almost the same structure with the 182-193 loop with AChBP, we compare two structures: <scene name='68/688431/Btx_complex_with_two_subunits/5'>removing</scene> the HAP form the structure and <scene name='68/688431/Btx_complex_with_two_subunits/6'> | ||
Revision as of 07:52, 2 February 2015
| |||||||||||
Quiz
References
- ↑ Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
- ↑ Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2013 Oct 28. PMID:24167270 doi:http://dx.doi.org/10.1073/pnas.1313156110
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009 Jan 1;457(7225):111-4. Epub 2008 Nov 5. PMID:18987633 doi:10.1038/nature07462
- ↑ Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor
- ↑ Samson AO, Levitt M. Inhibition mechanism of the acetylcholine receptor by alpha-neurotoxins as revealed by normal-mode dynamics. Biochemistry. 2008 Apr 1;47(13):4065-70. doi: 10.1021/bi702272j. Epub 2008 Mar 8. PMID:18327915 doi:http://dx.doi.org/10.1021/bi702272j
Proteopedia Page Contributors and Editors (what is this?)
Ma Zhuang, Zicheng Ye, Angel Herraez, Alexander Berchansky, Michal Harel