Binding site of AChR
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
There is a 13 amino acids high affinity peptide(<scene name='68/688431/Hap/2'>HAP</scene>) which corresponding to residues 187-199 of the AChR that can inhibits the binding of α-BTX to AChR. And the high affinity and specific interaction of α-bungarotoxin (<scene name='68/688431/Structure_of_btx/1'>α-BTX</scene>) with AChR has been of considerable importance in the study of the binding site of AChR.<ref name="Sussman 2001" >PMID:11683996</ref> The little peptide can bind to α-BTX as competitive inhibitors of α-BTX biding to AChR. So the complex between α-BTX and this little peptide(HAP) maybe can used as a model to study the binding site of AChR. | There is a 13 amino acids high affinity peptide(<scene name='68/688431/Hap/2'>HAP</scene>) which corresponding to residues 187-199 of the AChR that can inhibits the binding of α-BTX to AChR. And the high affinity and specific interaction of α-bungarotoxin (<scene name='68/688431/Structure_of_btx/1'>α-BTX</scene>) with AChR has been of considerable importance in the study of the binding site of AChR.<ref name="Sussman 2001" >PMID:11683996</ref> The little peptide can bind to α-BTX as competitive inhibitors of α-BTX biding to AChR. So the complex between α-BTX and this little peptide(HAP) maybe can used as a model to study the binding site of AChR. | ||
| - | The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail [http://en.wikipedia.org/wiki/Lymnaea_stagnalis Lymnaea stagnalis]. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref>PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref name="Sussman 2001" /> | + | The ligand binding site of AChR is mainly located at the α-subunits. The acetylcholine binding protein(<scene name='68/688431/Achbp/2'>AChBP</scene>) is most closely related to the α-subunits of the nAChR. AChBP is a soluble protein found in the snail [http://en.wikipedia.org/wiki/Lymnaea_stagnalis Lymnaea stagnalis]. Nearly all residues that are conserved within the nAChR family are present in AChBP, including those that are relevant for lignad binding.<ref name="AChBP">PMID:11357122</ref> And AChBP can also bind with α-Neurotoxins. So the AChBP structure is obviously an ideal candidate for testing the relevance of the conformation of the HAP when bound to α-BTX, to that of the corresponding binding region in AChR.<ref name="Sussman 2001" /> |
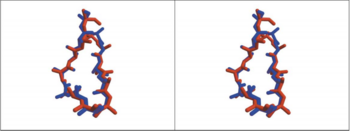
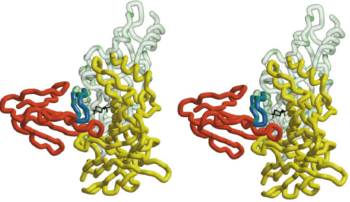
[[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. 4. Comparison, in Stere, of the 3D Structure of HAP(Red) and Loop 182-193 of AChBP(Blue)]] | [[Image:Comparison between HAP and AChBP.PNG|thumb|350px|Fig. 4. Comparison, in Stere, of the 3D Structure of HAP(Red) and Loop 182-193 of AChBP(Blue)]] | ||
| Line 43: | Line 43: | ||
The 13-mer <scene name='68/688431/Hap/2'>HAP</scene> assumes an antiparallel β hairpin structure, which can be used as a model to study the binding site of AChR. It is held snugly between <scene name='68/688431/Figure_1234/3'>fingers 1,2 and 4</scene> of α-BTX. The shortest and most numerous interactions are formed with <scene name='68/688431/Figure_1234/2'>finger 2</scene> of α-BTX. The two arms of the HAP hairpin assume a β sheet conformation, with residues Leu2 (corresponding to position 188 in AChR)-Tyr4 (corresponding to position 190 in AChR ) making an <scene name='68/688431/Residues_between_btx_and_hap/4'> intermolecular interaction </scene> with α-BTX residues Val39-Glu41 on a loop region. Tyr3 (corresponding to position 189 in AChr) of HAP forms a sung fit into a loop region of α-BTX. The formation of <scene name='68/688431/H_bond_between_hap_and_btx/2'>two H bonds</scene> from its hydroxyl to residues Thr8 and lle11 of α-BTX makes the tyrosine at that position an ideal candidate for forming binding interactions with α-BTX. Indeed, this tyrosine is known to play a crucial role in α-BTX binding. <ref name="Sussman 2001" /> | The 13-mer <scene name='68/688431/Hap/2'>HAP</scene> assumes an antiparallel β hairpin structure, which can be used as a model to study the binding site of AChR. It is held snugly between <scene name='68/688431/Figure_1234/3'>fingers 1,2 and 4</scene> of α-BTX. The shortest and most numerous interactions are formed with <scene name='68/688431/Figure_1234/2'>finger 2</scene> of α-BTX. The two arms of the HAP hairpin assume a β sheet conformation, with residues Leu2 (corresponding to position 188 in AChR)-Tyr4 (corresponding to position 190 in AChR ) making an <scene name='68/688431/Residues_between_btx_and_hap/4'> intermolecular interaction </scene> with α-BTX residues Val39-Glu41 on a loop region. Tyr3 (corresponding to position 189 in AChr) of HAP forms a sung fit into a loop region of α-BTX. The formation of <scene name='68/688431/H_bond_between_hap_and_btx/2'>two H bonds</scene> from its hydroxyl to residues Thr8 and lle11 of α-BTX makes the tyrosine at that position an ideal candidate for forming binding interactions with α-BTX. Indeed, this tyrosine is known to play a crucial role in α-BTX binding. <ref name="Sussman 2001" /> | ||
| - | In nAChR, the ligand-binding site is located at the interface between two subunits. The homopentameric α7 receptor contains five identical ligand binding sites. In these sites acrtylcholine is expected to bind through [http://en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interactions], where the positive charge of the quaternary ammonium of acetylcholine interacts with the electron-rich aromatic side chains.<ref | + | In nAChR, the ligand-binding site is located at the interface between two subunits. The homopentameric α7 receptor contains five identical ligand binding sites. In these sites acrtylcholine is expected to bind through [http://en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation-π interactions], where the positive charge of the quaternary ammonium of acetylcholine interacts with the electron-rich aromatic side chains.<ref name="AChBP" /> The ACh binding site in AChBP was assigned by the localization of a solvent molecule (positively charge HEPES) seen near residues corresponding to the 187-199 loop of the AChR α subunit and stacking on the corresponding Trp 143.<ref name="Sussman 2001" /> <scene name='68/688431/Hepes_five_subunits/2'>HEPES</scene> can be refined in the current AChBP structure, it does not make any specific hydrogen bonds with the protein, it stacks with its quaternary ammonium onto <scene name='68/688431/Hepes_trp143/1'>Trp 143</scene> making cation-π interactions as expected for nicotinic agonists.<ref name="AChBP" /> |
The superimposed model of AChBP and α-BTX suggests that the putative agonist HEPES seen in the AChBP structure is blocked from entering or leaving the AChBP interface cleft by the insertion of <scene name='68/688431/Hepes_black_loop_2/1'>loop 2</scene> of α-BTX into that cleft. This clarifies and explains the strong inhibition of AChR function by the toxin.<ref name="Sussman 2001" /> The superposition of the HAP on loop 182-193 of AChBP(Fig.5)show that the major interaction between α-BTX and AChR α subunit, occur in residues187-192 of that sununit. | The superimposed model of AChBP and α-BTX suggests that the putative agonist HEPES seen in the AChBP structure is blocked from entering or leaving the AChBP interface cleft by the insertion of <scene name='68/688431/Hepes_black_loop_2/1'>loop 2</scene> of α-BTX into that cleft. This clarifies and explains the strong inhibition of AChR function by the toxin.<ref name="Sussman 2001" /> The superposition of the HAP on loop 182-193 of AChBP(Fig.5)show that the major interaction between α-BTX and AChR α subunit, occur in residues187-192 of that sununit. | ||
Revision as of 14:38, 2 February 2015
| |||||||||||
Quiz
References
- ↑ Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
- ↑ Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2013 Oct 28. PMID:24167270 doi:http://dx.doi.org/10.1073/pnas.1313156110
- ↑ 3.0 3.1 3.2 3.3 Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009 Jan 1;457(7225):111-4. Epub 2008 Nov 5. PMID:18987633 doi:10.1038/nature07462
- ↑ 4.0 4.1 4.2 4.3 4.4 Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ 5.0 5.1 5.2 Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor
- ↑ Samson AO, Levitt M. Inhibition mechanism of the acetylcholine receptor by alpha-neurotoxins as revealed by normal-mode dynamics. Biochemistry. 2008 Apr 1;47(13):4065-70. doi: 10.1021/bi702272j. Epub 2008 Mar 8. PMID:18327915 doi:http://dx.doi.org/10.1021/bi702272j
Proteopedia Page Contributors and Editors (what is this?)
Ma Zhuang, Zicheng Ye, Angel Herraez, Alexander Berchansky, Michal Harel