Binding site of AChR
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
[[Image:Mechanism of GLIC.PNG|thumb|350px|Fig. 3. Open GLIC and closed ELIC structure comarison green is GLIC and red is ELIC]] | [[Image:Mechanism of GLIC.PNG|thumb|350px|Fig. 3. Open GLIC and closed ELIC structure comarison green is GLIC and red is ELIC]] | ||
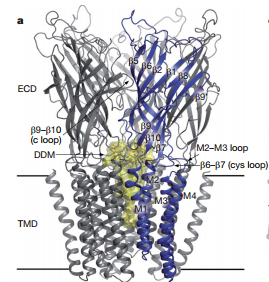
| - | The general mechanism of pLGIC is provided by Prof. Jean-Pierre Changeux in the paper 'X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation[J]. Nature, 2009, 457(7225): 111-114'. GLIC and ELIC are both pentameric ligand gated ion channel which in the same family with AChR and AChBP. GLIC is an apparently open conformation while | + | The general mechanism of pLGIC is provided by Prof. Jean-Pierre Changeux in the paper 'X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation[J]. Nature, 2009, 457(7225): 111-114'. GLIC and ELIC are both pentameric ligand gated ion channel which in the same family with AChR and AChBP. GLIC is an apparently open conformation while ELIC is presumed closed conformation. Comparative analysis of GLIC and ELIC reveals the rotation of β-sandwich and a tilt of M2 and M3, which will show the mechanism of pLGIC opening. |
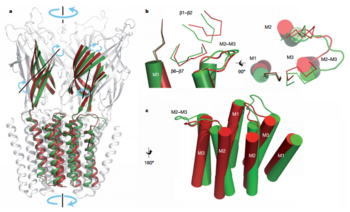
| - | But in the first 100 lowest-frequency modes only 50% of the transition can be explained. The rest and more local movements occur: in the TMD the outer ends of M2 and M3 of GLIC are tilted away radially from the channel axis, while the outer end of M1 is fixed. The inner ends of M1, M2 and M3 move tangentially towards the left, when viewed from the membrane (Fig. 3b). In the ECD, the core of the β-sandwich undergoes little deformation, but is rotated by 8° around an axis roughly perpendicular to the inner sheet of the β-sandwich (Fig. 3a), concomitant with a rearrangement of both the subunit–subunit and the ECD/TMD interfaces, regions known to contribute to neurotransmitter gating. A downward motion of the β1–β2 loop, concomitant with a displacement of the M2–M3 loop, M2 and M3 helices and β6–β7 loop towards the periphery of the molecule (Fig. 3c), thereby opening the pore<ref name="Bocquet2009" />. | + | |
| + | Common core is consists by M1, M2 and M3, and a large portion of the β-sandwich. The superimposition of common core shows that the GLIC subunits display an anticlock quaternary twist compared to ELIC(Fig. 3a). But in the first 100 lowest-frequency modes only 50% of the transition can be explained. The rest and more local movements occur: in the TMD the outer ends of M2 and M3 of GLIC are tilted away radially from the channel axis, while the outer end of M1 is fixed. The inner ends of M1, M2 and M3 move tangentially towards the left, when viewed from the membrane (Fig. 3b). In the ECD, the core of the β-sandwich undergoes little deformation, but is rotated by 8° around an axis roughly perpendicular to the inner sheet of the β-sandwich (Fig. 3a), concomitant with a rearrangement of both the subunit–subunit and the ECD/TMD interfaces, regions known to contribute to neurotransmitter gating. A downward motion of the β1–β2 loop, concomitant with a displacement of the M2–M3 loop, M2 and M3 helices and β6–β7 loop towards the periphery of the molecule (Fig. 3c), thereby opening the pore<ref name="Bocquet2009" />. | ||
At the beginning, this kind of twist to open motions is come from ''ab initio'' normal mode analysis of nAChRs, and then plausibly be extended to the pLGICs family. The structural transition described here couples in an allosteric manner the opening–closing motion of the pore with distant binding sites—located at the ECD subunit interface for neurotransmitters, or within the TMD for allosteric effectors30—and may possibly serve as a general mechanism of signal transduction in pLGICs<ref name="Bocquet2009" />. | At the beginning, this kind of twist to open motions is come from ''ab initio'' normal mode analysis of nAChRs, and then plausibly be extended to the pLGICs family. The structural transition described here couples in an allosteric manner the opening–closing motion of the pore with distant binding sites—located at the ECD subunit interface for neurotransmitters, or within the TMD for allosteric effectors30—and may possibly serve as a general mechanism of signal transduction in pLGICs<ref name="Bocquet2009" />. | ||
Revision as of 15:31, 2 February 2015
| |||||||||||
Quiz
References
- ↑ Purves, Dale, George J. Augustine, David Fitzpatrick, William C. Hall, Anthony-Samuel LaMantia, James O. McNamara, and Leonard E. White (2008). Neuroscience. 4th ed. Sinauer Associates. pp. 156–7. ISBN 978-0-87893-697-7.
- ↑ Gonzalez-Gutierrez G, Cuello LG, Nair SK, Grosman C. Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2013 Oct 28. PMID:24167270 doi:http://dx.doi.org/10.1073/pnas.1313156110
- ↑ 3.0 3.1 3.2 3.3 3.4 Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009 Jan 1;457(7225):111-4. Epub 2008 Nov 5. PMID:18987633 doi:10.1038/nature07462
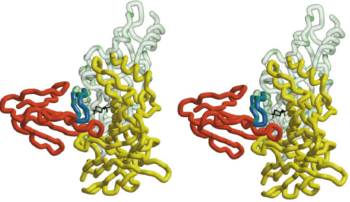
- ↑ 4.0 4.1 4.2 4.3 4.4 Harel M, Kasher R, Nicolas A, Guss JM, Balass M, Fridkin M, Smit AB, Brejc K, Sixma TK, Katchalski-Katzir E, Sussman JL, Fuchs S. The binding site of acetylcholine receptor as visualized in the X-Ray structure of a complex between alpha-bungarotoxin and a mimotope peptide. Neuron. 2001 Oct 25;32(2):265-75. PMID:11683996
- ↑ 5.0 5.1 5.2 Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001 May 17;411(6835):269-76. PMID:11357122 doi:10.1038/35077011
- ↑ http://en.wikipedia.org/wiki/Nicotinic_acetylcholine_receptor
- ↑ Samson AO, Levitt M. Inhibition mechanism of the acetylcholine receptor by alpha-neurotoxins as revealed by normal-mode dynamics. Biochemistry. 2008 Apr 1;47(13):4065-70. doi: 10.1021/bi702272j. Epub 2008 Mar 8. PMID:18327915 doi:http://dx.doi.org/10.1021/bi702272j
Proteopedia Page Contributors and Editors (what is this?)
Ma Zhuang, Zicheng Ye, Angel Herraez, Alexander Berchansky, Michal Harel