This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Ubc9
From Proteopedia
| Line 3: | Line 3: | ||
== Structure == | == Structure == | ||
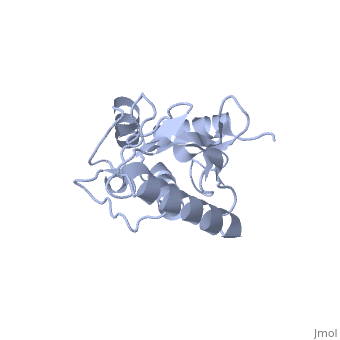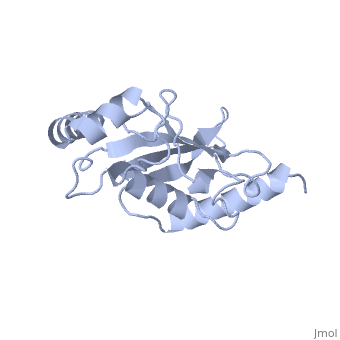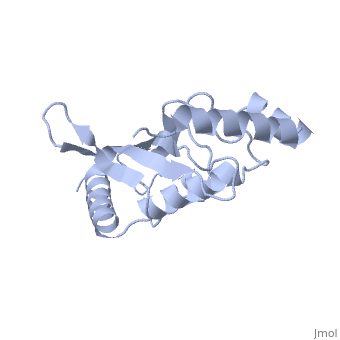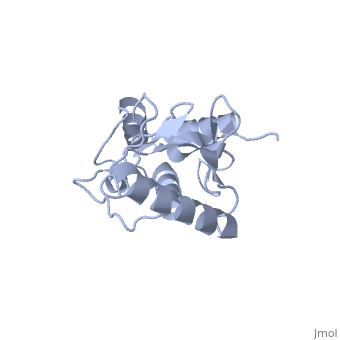
| + | Ubc9 exhibits a single domain structure consisting of both alpha helices and beta-pleated sheets. C93 has been identified as the active residue and is located on a non-secondary structured loop of amino acids (78-108) ^1^ . | ||
== Function == | == Function == | ||
| Line 17: | Line 18: | ||
== Structural highlights == | == Structural highlights == | ||
| - | Anything in this section will appear adjacent to the 3D structure and will be scrollable. | ||
== References == | == References == | ||
| + | 1. Reverter, D., & Lima, C. D. (2005). Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature, 435(June), 687–692. doi:10.1038/nature03588 | ||
<references/> | <references/> | ||
Revision as of 04:50, 22 February 2015
|
Contents |
Structure
Ubc9 exhibits a single domain structure consisting of both alpha helices and beta-pleated sheets. C93 has been identified as the active residue and is located on a non-secondary structured loop of amino acids (78-108) ^1^ .
Function
test
Disease
test
Relevance
test
Structural highlights
References
1. Reverter, D., & Lima, C. D. (2005). Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature, 435(June), 687–692. doi:10.1038/nature03588