Ubc9
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
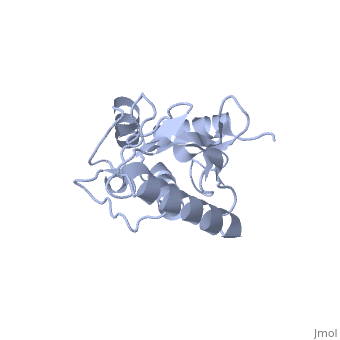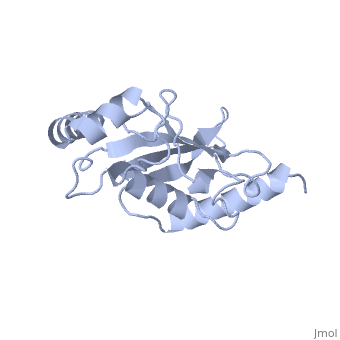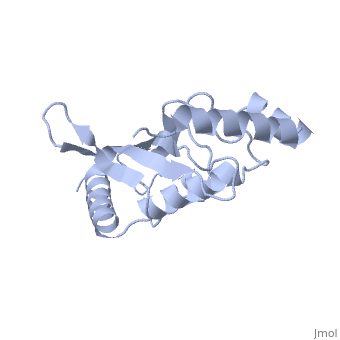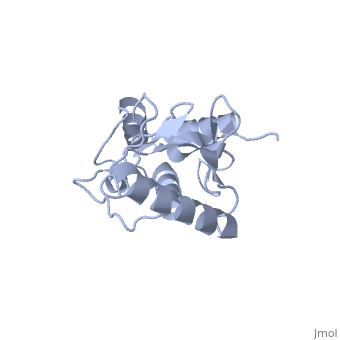
''Ubc9'' exhibits a single domain structure consisting of both alpha helices and beta-pleated sheets. Cys93 has been identified as the active residue and is located on a non-secondary structured loop of amino acids (78-108) between beta sheet four and alpha helix two ^1^ . | ''Ubc9'' exhibits a single domain structure consisting of both alpha helices and beta-pleated sheets. Cys93 has been identified as the active residue and is located on a non-secondary structured loop of amino acids (78-108) between beta sheet four and alpha helix two ^1^ . | ||
| + | |||
| + | <Structure load='2UYZ' size='350' frame='true' align='left' caption='Noncovalent Ubc9 and SUMO1 complex' scene='Insert optional scene name here' /> | ||
== Function == | == Function == | ||
Revision as of 03:17, 24 February 2015
| |||||||||||
References
1. Reverter, D., & Lima, C. D. (2005). Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature, 435(June), 687–692. doi:10.1038/nature03588