This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Z-DNA
From Proteopedia
| Line 15: | Line 15: | ||
ADAR1 (<scene name='Sandbox_Z-DNA/Adar1/3'>Z-ALPHA and Z-DNA complex</scene>, [[1qbj]]) belongs to the family of deaminases that modify double stranded mRNA by catalyzing the conversion of adenine to inosine which is then translated to guanosine. It is a complex protein with two Z-DNA binding motifs called <scene name='Sandbox_Z-DNA/Adar1zalpha/10'>Z-alpha</scene> and Z-beta.<ref name = 'Wang'>PMID:17485386</ref> ADAR1 also has three copies of double-stranded RNA binding motif (DRBM) and a catalytic domain related to ''E.coli'' cytidine deaminase. The binding motif Z-alpha belongs to winged-helix-turn-helix family of proteins. It consists of a <scene name='Z-DNA/Adar1zalpha/1'>helix-turn-helix motif</scene> which has two alpha helices (<scene name='Z-DNA/Adar1zalpha/2'>alpha-2</scene> and <scene name='Z-DNA/Adar1zalpha/3'>alpha-3 also called the recognition</scene>) connected by a short strand of amino acids and a <scene name='Sandbox_Z-DNA/Adar1zalpha/15'>C- terminal beta-sheet</scene>. The beta sheet constrains the fold by contacting the residues between alpha-2 and alpha-3. | ADAR1 (<scene name='Sandbox_Z-DNA/Adar1/3'>Z-ALPHA and Z-DNA complex</scene>, [[1qbj]]) belongs to the family of deaminases that modify double stranded mRNA by catalyzing the conversion of adenine to inosine which is then translated to guanosine. It is a complex protein with two Z-DNA binding motifs called <scene name='Sandbox_Z-DNA/Adar1zalpha/10'>Z-alpha</scene> and Z-beta.<ref name = 'Wang'>PMID:17485386</ref> ADAR1 also has three copies of double-stranded RNA binding motif (DRBM) and a catalytic domain related to ''E.coli'' cytidine deaminase. The binding motif Z-alpha belongs to winged-helix-turn-helix family of proteins. It consists of a <scene name='Z-DNA/Adar1zalpha/1'>helix-turn-helix motif</scene> which has two alpha helices (<scene name='Z-DNA/Adar1zalpha/2'>alpha-2</scene> and <scene name='Z-DNA/Adar1zalpha/3'>alpha-3 also called the recognition</scene>) connected by a short strand of amino acids and a <scene name='Sandbox_Z-DNA/Adar1zalpha/15'>C- terminal beta-sheet</scene>. The beta sheet constrains the fold by contacting the residues between alpha-2 and alpha-3. | ||
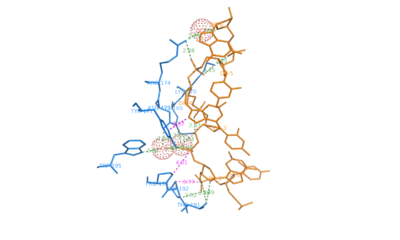
| - | The contact surface between <scene name='Sandbox_Z-DNA/Adar1/4'>Z-alpha and DNA</scene> consists of residues from the helix alpha-3 and COOH-terminal beta hairpin. Hydrogen bonding is present between <scene name='Z-DNA/Aminoacid/1'>amino acids</scene> Lys<sup>169</sup>, Lys <sup>170</sup>, Asn<sup>173</sup>, Arg<sup>174</sup> and Tyr<sup>177</sup> in the helix alpha-3 and <scene name='Z-DNA/Dnanucleotides/2'>five consecutive phosphates on Z-DNA</scene>. Lys<sup>169</sup>, Asn<sup>173</sup>, Arg<sup>174</sup>, Trp<sup>195</sup> make water mediated phosphate contacts with Z-DNA. In addition Thr<sup>191</sup> and Arg<sup>174</sup> <scene name='Z-DNA/Thrarg/1'>bind to the furanose oxygens</scene> of G2 and G6 on Z-DNA. An important interaction is the <scene name='Z-DNA/Tyrosine_and_g4/1'>Vanderwaal's bond</scene> between aromatic ring of Tyr<sup>177</sup> and the carbon 8 of G4. This is unique to Z-DNA as the interaction requires the base to be in syn conformation. Pro<sup>192</sup>, Pro <sup>193</sup> form another set of <scene name='Z-DNA/Pro/1'>important Vanderwaal's interactions</scene> with Z-DNA where the pyrrolidine rings bond with the sugar-phosphate backbone from phosphate 2 to phosphate 3. Pro<sup>192</sup> is conserved in Z-alpha and its homologues and forms a cis peptide bond which positions beta loop against the Z-DNA surface.<ref name = SchwartzRich>PMID: 10364558</ref> | + | The contact surface between <scene name='Sandbox_Z-DNA/Adar1/4'>Z-alpha and DNA</scene> consists of residues from the helix alpha-3 and COOH-terminal beta hairpin. Hydrogen bonding is present between <scene name='Z-DNA/Aminoacid/1'>amino acids</scene> Lys<sup>169</sup>, Lys <sup>170</sup>, Asn<sup>173</sup>, Arg<sup>174</sup> and Tyr<sup>177</sup> in the helix alpha-3 and <scene name='Z-DNA/Dnanucleotides/2'>five consecutive phosphates on Z-DNA</scene>. Lys<sup>169</sup>, Asn<sup>173</sup>, Arg<sup>174</sup>, Trp<sup>195</sup> make water mediated phosphate contacts with Z-DNA. In addition Thr<sup>191</sup> and Arg<sup>174</sup> <scene name='Z-DNA/Thrarg/1'>bind to the furanose oxygens</scene> of G2 and G6 on Z-DNA. An important interaction is the <scene name='Z-DNA/Tyrosine_and_g4/1'>Vanderwaal's bond</scene> between aromatic ring of Tyr<sup>177</sup> and the carbon 8 of G4. This is unique to Z-DNA as the interaction requires the base to be in syn conformation. Pro<sup>192</sup>, Pro <sup>193</sup> form another set of <scene name='Z-DNA/Pro/1'>important Vanderwaal's interactions</scene> with Z-DNA where the pyrrolidine rings bond with the sugar-phosphate backbone from phosphate 2 to phosphate 3. Pro<sup>192</sup> is conserved in Z-alpha and its homologues and forms a cis peptide bond which positions beta loop against the Z-DNA surface.<ref name = SchwartzRich>PMID: 10364558</ref> |
| + | |||
[[Image:Hbonding_Z-DNA.png|left|thumb|400px|Polar Interactions between ADAR1 and Z-DNA]] | [[Image:Hbonding_Z-DNA.png|left|thumb|400px|Polar Interactions between ADAR1 and Z-DNA]] | ||
| - | + | {{Clear}} | |
The double stranded RNA substrate for ADAR1 is formed by folding of 3' intron back onto the exon containing the site to be edited. This shows that the editing of RNA occurs before the splicing of RNA providing an explanation for the binding of Z-DNA by ADAR1. Z-DNA may localize the editing activity of ADAR1 to a particular region within a gene, thus preventing indiscriminate modification. This allows for editing of the nascent transcript and blocks further transcription of gene. It has also been suggested that the extent of adenosine to inosine is proportional to amount of Z-DNA and also the ease with which the surrounding sequences adopt Z-DNA conformation. According to a study binding of ADAR1 to Z-DNA resulted in the increase in promoter activity of the gene which suggests that Z-DNA formation in the promoter region is itself involved in the regulation of transcription.<ref name = 'Rich'>PMID:12838348</ref> | The double stranded RNA substrate for ADAR1 is formed by folding of 3' intron back onto the exon containing the site to be edited. This shows that the editing of RNA occurs before the splicing of RNA providing an explanation for the binding of Z-DNA by ADAR1. Z-DNA may localize the editing activity of ADAR1 to a particular region within a gene, thus preventing indiscriminate modification. This allows for editing of the nascent transcript and blocks further transcription of gene. It has also been suggested that the extent of adenosine to inosine is proportional to amount of Z-DNA and also the ease with which the surrounding sequences adopt Z-DNA conformation. According to a study binding of ADAR1 to Z-DNA resulted in the increase in promoter activity of the gene which suggests that Z-DNA formation in the promoter region is itself involved in the regulation of transcription.<ref name = 'Rich'>PMID:12838348</ref> | ||
Revision as of 12:48, 25 February 2015
| |||||||||||
Contents |
Movie Depicting ADAR1 binding to Z-DNA
Comparison of the three helices and helical parameters of DNA
Sources[6]
|
|
|
| Parameter | A-DNA | B-DNA | Z-DNA |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Residues per turn | 11 | 10.5 | 12 |
| Axial rise [Å] | 2.55 | 3.4 | 3.7 |
| Helix pitch(°) | 28 | 34 | 45 |
| Base pair tilt(°) | 20 | −6 | 7 |
| Rotation per residue (°) | 33 | 36 | -30 |
| Diameter of helix [Å] | 23 | 20 | 18 |
| Glycosidic bond configuration dA,dT,dC dG | anti anti | anti anti | anti syn |
| Sugar pucker dA,dT,dC dG | C3'-endo C3'-endo | C2'-endo C2'-endo | C2'-endo C3'-endo |
| Intrastrand phosphate-phosphate distance [Å] dA,dT,dC dG | 5.9 5.9 | 7.0 7.0 | 7.0 5.9 |
| Sources:[7][8][9] | |||
3D structures of Z-DNA
Updated on 25-February-2015
1woe, 3qba – ZDNA hexamer CGCGCG
1qbj – ZDNA hexamer CGCGCG + hADAR1 Zα domain - human
2heo - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zα domain – mouse
2eyi - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zβ domain
1j75 - ZDNA hexamer CGCGCG + DLM-1 Zα domain
Additional Resources
For additional information, see: Nucleic Acids
References
- ↑ 1.0 1.1 1.2 Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003 Jul;4(7):566-72. PMID:12838348 doi:10.1038/nrg1115
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007 May 1;12:4424-38. PMID:17485386
- ↑ Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005 Oct 20;437(7062):1183-6. PMID:16237447 doi:10.1038/nature04088
- ↑ Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999 Jun 11;284(5421):1841-5. PMID:10364558
- ↑ Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1514-8. Epub 2004 Feb 2. PMID:14757814 doi:10.1073/pnas.0308260100
- ↑ http://203.129.231.23/indira/nacc/
- ↑ Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791-846. PMID:6383204 doi:http://dx.doi.org/10.1146/annurev.bi.53.070184.004043
- ↑ Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680-6. PMID:514347
- ↑ Sinden, Richard R (1994-01-15). DNA structure and function (1st ed.). Academic Press. pp. 398. ISBN 0-12-645750-6.
Proteopedia Page Contributors and Editors (what is this?)
Adithya Sagar, Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Donald Voet, Jaime Prilusky, Karl Oberholser, Eran Hodis