UBC13 MMS2
From Proteopedia
| Line 30: | Line 30: | ||
Ubc13 weights 17.6 kDA, and Mms2 is 16.8 kDA<ref name=Pastushok> Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. The heterodimer is stable at high stalt concentrations (1 M), suggesting strong interactions between the two. Kd between the Ubc13 and Mms2 is 2 uM. | Ubc13 weights 17.6 kDA, and Mms2 is 16.8 kDA<ref name=Pastushok> Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. The heterodimer is stable at high stalt concentrations (1 M), suggesting strong interactions between the two. Kd between the Ubc13 and Mms2 is 2 uM. | ||
| - | <scene name='69/695700/Ubc13_mms2_phe57/1'>Phe57</scene> and Glu55 of Ubc13 interact with the N-terminal domain of Mms2 to ensure stable docking<ref name=Pastushok> Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. Additionally, Arg70 hydrophobically interacts with an alpha helix of Mms2 in two places<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. Mms2’s Phe13 is inserted between Glu55, Phe57, and Arg70 of Ubc13 to create a hydrophobic pocket<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. It is therorized that Glu55 and Arg70 of Ubc13 are more important for recognition instead of stability<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. | + | <scene name='69/695700/Ubc13_mms2_phe57/1'>Phe57</scene> and <scene name='69/695700/Ubc13_mms2_glu55/1'>Glu55</scene> of Ubc13 interact with the N-terminal domain of Mms2 to ensure stable docking<ref name=Pastushok> Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. Additionally, Arg70 hydrophobically interacts with an alpha helix of Mms2 in two places<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. Mms2’s Phe13 is inserted between Glu55, Phe57, and Arg70 of Ubc13 to create a hydrophobic pocket<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. It is therorized that Glu55 and Arg70 of Ubc13 are more important for recognition instead of stability<ref name=Pastushok> 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.</ref>. |
==Mechanism== | ==Mechanism== | ||
Revision as of 02:28, 26 February 2015
|
Contents |
Summary
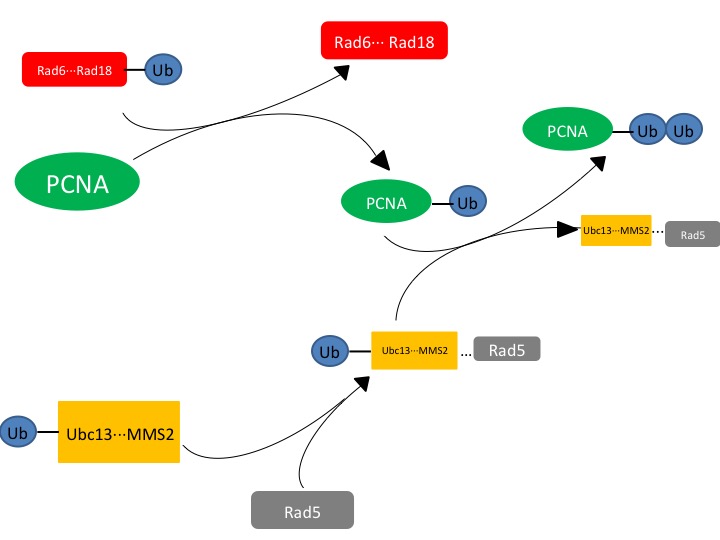
Ubc13 is an E2 ubiquitin-conjugating enzyme that can form a heterodimer with Mms2 to function as a part of the translesion synthesis (TLS) pathway,[1][2]. When bound to Mms2, Ubc13 will polyubiquitinate proliferating cell nuclear antigen (PCNA), a sliding clamp protein at the DNA transcription fork[1]. Ubc13-Mms2 functions to polyubiquitinate PCNA following the initial monoubiquitination by Rad6-Rad18 (another E2 complex)[2].
Function
Ubc13 functions as a heterodimer with Mms2, a structurally similar protein to Ubc13 that lacks the catalytic cysteine residue in the active site[3][4]. The Ubc13-E2 complex with Mms2 functions primarily to enhance DNA repair from double stranded breaks. Mms2 bound to Ubc13 helps orient the ubiquitin molecule for proper ubiquitination of the K63 residue[5]. Mms2 is considered a Ubiquitin E2 variant (UEV) protein, because it lacks the catalytic cysteine residue necessary for proper thioester formation[5][6][3].
Regulation
The regulation of Ubc13 is controlled by the competitive binding of the two different UEV's, Mms2 and UEV1A[2]. Ubc13 binding to Mms2 activates the DNA repair pathway, while Ubc13 binding to UEV1A activates the NF-kappaB pathway, a gene regulation pathway involved in DNA transcription factors[2]. These two opposing pathways being downstream targets for Ubc13-bound complexes is the basis for regulation.
Coordinating Enzymes
- Another E2 complex, Rad6-Rad18, starts the process of DNA repair by monoubiquitinating PCNA near the replication fork of DNA. This DNA repair will arrest cell cycle progression until DNA repair is complete[2].
- UEV1A, another cofactor enzyme that binds to Ubc13, is thought to compete with Mms2 for binding to Ubc13. This is thought to be a regulatory mechanism for Ubc13 activity in the nucleus of cells[2].
- Several DNA polymerases such as rev1, pol eta, and pol zeta contain Ubiquitin-binding domains that recognize K164 polyubiquitination of PCNA[1].
- Rad5, an E3 RING (Really Interesting New Gene) protein, interacts with the Ubc13-Mms2 heterodimer in order to ligate the ubiquitin on the PCNA. Rad5, as well as Rad18 (RING proteins) are involved in the recruitment of Ubc13-Mms2 heterodimer formation[2].
Pathway for DNA Repair
Figure. 1. The affected protein PCNA is a DNA clamp that is monoubiquitinated by the Rad6-Rad18 complex. The monoubiquitinated PCNA is then polyubuiquitinated by the UBC13-Mms2-Rad5 complex.
Structural Highlights/Important Residues
Ubc13 weights 17.6 kDA, and Mms2 is 16.8 kDA[4]. The heterodimer is stable at high stalt concentrations (1 M), suggesting strong interactions between the two. Kd between the Ubc13 and Mms2 is 2 uM. and of Ubc13 interact with the N-terminal domain of Mms2 to ensure stable docking[4]. Additionally, Arg70 hydrophobically interacts with an alpha helix of Mms2 in two places[4]. Mms2’s Phe13 is inserted between Glu55, Phe57, and Arg70 of Ubc13 to create a hydrophobic pocket[4]. It is therorized that Glu55 and Arg70 of Ubc13 are more important for recognition instead of stability[4].
Mechanism
The exact mechanism for how Ubc13 transfers ubiquitin is not known, however the mechanism occurs in either a step-wise or concerted reaction.
References
- ↑ 1.0 1.1 1.2 3. Halas, A.; Podlaska, A. F.; Derkacz, J. F.; McIntyre, J. F.; Skoneczna, A. F.; Sledziewska-Gojska, E. The roles of PCNA SUMOylation, Mms2-Ubc13 and Rad5 in translesion DNA synthesis in Saccharomyces cerevisiae. Molecular microbiology JID - 8712028 0809.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 1. Andersen, P. L.; Zhou, H. F.; Pastushok, L. F.; Moraes, T. F.; McKenna, S. F.; Ziola B FAU - Ellison, Michael,J.; FAU, E. M.; FAU, D. V.; Xiao, W. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. The Journal of cell biology JID - 0375356 1107.
- ↑ 3.0 3.1 5. McKenna, S.; Spyracopoulos, L. F.; Moraes, T. F.; Pastushok, L. F.; Ptak, C. F.; Xiao W FAU - Ellison,,M.J.; Ellison, M. J. Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. The Journal of biological chemistry JID - 2985121R 1207.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 7. Pastushok, L.; FAU, M. T.; FAU, E. M.; Xiao, W. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. The Journal of biological chemistry JID - 2985121R 0902.
- ↑ 5.0 5.1 9. VanDemark, A. P.; FAU, H. R.; Tsui C FAU - Pickart,,C.M.; FAU, P. C.; Wolberger, C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell JID - 0413066 0726.
- ↑ 6. Moraes, T. F.; FAU, E. R.; McKenna, S. F.; Pastushok, L. F.; Xiao W FAU - Glover,,J.N.; FAU, G. J.; Ellison, M. J. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nature structural biology JID - 9421566 0816.
Proteopedia Page Contributors and Editors (what is this?)
David A Taves, Michal Harel, Christopher Alexander Hudson, Nicholas R. Dunham