AChE inhibitors and substrates
From Proteopedia
(Difference between revisions)
| Line 36: | Line 36: | ||
====DFP==== | ====DFP==== | ||
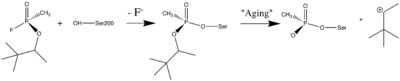
<scene name='2dfp/Cv/3'>DFP</scene>, di''iso''propylphosphorofluoridate, is an other toxic OP nerve agent. It is also inhibits AChE by covalent binding to the catalytic Ser200. As in the case with soman ([[1som]]) and sarin ([[1cfj]]), there are four hydrogen bond donors (dotted lines) to the anionic phosphonyl oxygen atoms: the backbone amide nitrogen atoms of Ala201, Gly118, and Gly119, as well as His440 Nε2 at the <scene name='2dfp/Cv/4'>active site</scene> of aged DFP-TcAChE ([[2dfp]]). Phosphorylation with DFP caused an unexpected distortion in the main chain of a loop that includes residues F288 and F290 of the TcAChE acyl binding pocket. F288 and F290 move significantly in the <span style="color:lime;background-color:black;font-weight:bold;">DFP-TcAChE structure (green)</span>, in comparison to their positions in the <font color='magenta'><b>native enzyme</b></font> ([[2ace]]). This is the first major conformational change reported in the active site of any AChE−ligand complex, and it offers a structural explanation for the substrate selectivity of AChE <ref name="Millard"/>. | <scene name='2dfp/Cv/3'>DFP</scene>, di''iso''propylphosphorofluoridate, is an other toxic OP nerve agent. It is also inhibits AChE by covalent binding to the catalytic Ser200. As in the case with soman ([[1som]]) and sarin ([[1cfj]]), there are four hydrogen bond donors (dotted lines) to the anionic phosphonyl oxygen atoms: the backbone amide nitrogen atoms of Ala201, Gly118, and Gly119, as well as His440 Nε2 at the <scene name='2dfp/Cv/4'>active site</scene> of aged DFP-TcAChE ([[2dfp]]). Phosphorylation with DFP caused an unexpected distortion in the main chain of a loop that includes residues F288 and F290 of the TcAChE acyl binding pocket. F288 and F290 move significantly in the <span style="color:lime;background-color:black;font-weight:bold;">DFP-TcAChE structure (green)</span>, in comparison to their positions in the <font color='magenta'><b>native enzyme</b></font> ([[2ace]]). This is the first major conformational change reported in the active site of any AChE−ligand complex, and it offers a structural explanation for the substrate selectivity of AChE <ref name="Millard"/>. | ||
| + | |||
| + | {{Clear}} | ||
| + | ==AChE monovalent inhibitors (continuation of the page [[AChE inhibitors and substrates]])== | ||
| + | ===Treatment of Alzheimer's disease=== | ||
| + | {{Clear}} | ||
| + | [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease] (AD) is a disorder that attacks the [http://en.wikipedia.org/wiki/Central_nervous_system central nervous system] through progressive degeneration of its neurons. Patients with this disease develop [http://en.wikipedia.org/wiki/Dementia dementia] which becomes more severe as the disease progresses. It was suggested that symptoms of AD are caused by decrease of activity of [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Neocortex neocortical] and [http://en.wikipedia.org/wiki/Hippocampus hippocampal] neurons. Treatment of AD by ACh precursors and [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Agonist agonists] was ineffective or caused severe side effects. ACh hydrolysis by AChE causes termination of cholinergic neurotransmission. Therefore, compounds which inhibit AChE might significantly increase the levels of ACh depleted in AD. Indeed, it was shown that [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitors] improve the cognitive abilities of AD patients at early stages of the disease development. The first generation of AD drugs were AChE inhibitors: alcaloids like [http://en.wikipedia.org/wiki/Huperzine_A (-)-Huperzine A (HupA)] and [http://en.wikipedia.org/wiki/Galantamine (-)-galanthamine (GAL, Reminyl)]; [http://en.wikipedia.org/wiki/Chemical_synthesis synthetic] compounds [http://en.wikipedia.org/wiki/Tacrine tacrine (Cognex)] and [http://en.wikipedia.org/wiki/Rivastigmine rivastigmine (Exelon)]. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====(-)-Huperzine A==== | ||
| + | [http://en.wikipedia.org/wiki/Huperzine_A (-)-Huperzine A], discovered by Chinese scientists from 1980s, has been proved to be a powerful, highly specific, and [http://en.wikipedia.org/wiki/Enzyme_inhibitor#Reversible_inhibitors reversible inhibitor] of AChE. It is a novel [http://en.wikipedia.org/wiki/Alkaloid alkaloid] originally isolated from the '''Traditional Chinese medicine''' [http://en.wikipedia.org/wiki/Traditional_Chinese_medicine] Qian Ceng Ta which is produced from the whole plant of the [http://en.wikipedia.org/wiki/Firmoss firmoss][http://en.wikipedia.org/wiki/Huperzia_serrata ''Huperzia serrata'']. Qian Ceng Ta has been used for over 1000 years in China for treatment of [http://en.wikipedia.org/wiki/Bruise contusions], [http://en.wikipedia.org/wiki/Strain_(injury) strains], [http://en.wikipedia.org/wiki/Swelling_(medical) swellings], [http://en.wikipedia.org/wiki/Schizophrenia schizophrenia] and [http://en.wikipedia.org/wiki/Myasthenia_gravis myasthenia gravis]. Shuangyiping[http://www.54md.com/drugstore/pic/gpic_25fd25197010a0fb4a680516735e613c.jpg], a tablet form of HupA produced from the extracts of ''Huperzia serrata'', was developed in 1996 as a new drug for symptomatic treatment of Alzheimer’s disease in China. Compared with the other three [http://en.wikipedia.org/wiki/Food_and_Drug_Administration_(United_States) FDA]-approved drugs for the treatment of Alzheimer’s disease, Donepezil (Aricept), Rivastigmine (Exelon), Galanthamine (Reminyl), HupA has better penetration through the [http://en.wikipedia.org/wiki/Blood-brain_barrier blood-brain barrier], higher oral [http://en.wikipedia.org/wiki/Bioavailability bioavailability], and longer duration of AChE inhibitory action. The structure of HupA shows some similarity to other known AChE inhibitors. The molecule is fairly rigid and contains an [http://en.wikipedia.org/wiki/Aromaticity aromatic] system as well as a [http://en.wikipedia.org/wiki/Amine primary amino group] that is probably [http://en.wikipedia.org/wiki/Protonation protonated] at physiological [http://en.wikipedia.org/wiki/PH pH]. Various suggestions have been made with respect to its orientation within the active site of AChE, and with respect to the amino acid residue with which its putative [http://en.wikipedia.org/wiki/Pharmacophore pharmacophoric] groups might interact. Solution of the 3D structure of a complex of HupA with AChE would permit unequivocal resolution of this issue and it would also provide a rational basis for structure-related [http://en.wikipedia.org/wiki/Drug_design drug design] aimed at developing synthetic analogues of HupA with improved therapeutic properties. | ||
| + | |||
| + | The [http://en.wikipedia.org/wiki/X-ray_crystallography crystal structure] of the complex of ''Tc''AChE with HupA at 2.5 Å resolution ([[1vot]]) was determined in 1997 and it shows an unexpected orientation for the inhibitor with surprisingly few strong direct interactions with protein residues to explain its high affinity. <font color='blueviolet'><b>HupA</b></font> binds to ''Tc''AChE at the active site, and its <scene name='1vot/Active_site/8'>observed orientation is almost orthogonal</scene> in comparison to <font color='gray'><b>ACh</b></font>. The principal interactions of <scene name='1vot/1vot_ache_interactions/2'>HupA with TcAChE</scene> are including: a direct <scene name='1vot/1vot_199_130_117/2'>hydrogen bond with Tyr130 and HBs with Glu199 and Gly117</scene> <span style="color:orange;background-color:black;font-weight:bold;">(colored orange)</span> through a water molecule as a linker at the bottom of the gorge; [http://en.wikipedia.org/wiki/Cation-pi_interaction cation-π] interactions between the amino group of <scene name='1vot/1vot_84_330/2'>HupA and Trp84 and Phe330</scene> <span style="color:lime;background-color:black;font-weight:bold;">(colored green)</span> with the distance between the nitrogen and the centroid of the aromatic rings of 4.8 and 4.7 Å, respectively; at the top of the gorge, [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonds] through two water molecules as linkers formed between the amino group of <scene name='1vot/1vot_70_72_81_85_121/3'>HupA and Tyr70, Asp72, Ser81, Asn85 and Tyr121</scene> <font color='magenta'><b>(colored magenta)</b></font>. An unusually short (~3.0 Å) C-H→O HB has been seen between the ethylidene methyl group of <scene name='1vot/1vot_440/2'>HupA and the main chain oxygen of His440</scene> <font color='crimson'><b>(colored crimson)</b></font> <ref name="Raves">PMID:8989325</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | |||
| + | ====Tacrine==== | ||
| + | <scene name='AChE_inhibitors_and_substrates/Com_view_tacrine/1'>Tacrine</scene> [http://en.wikipedia.org/wiki/Tacrine]. In the X-ray crystal structure of ''Tc''AChE/<scene name='AChE_inhibitors_and_substrates/Com_view_tacrine/2'>tacrine</scene> complex which was determined at 2.8 Å resolution, the tacrine is seen <font color='magenta'><b>(magenta)</b></font> bound in the active site of ''Tc''AChE ([[1acj]]) <ref name="Harel">PMID:8415649</ref>. <font color='gray'><b>ACh (gray)</b></font> is shown for comparison. In the crystal structure of ''[http://en.wikipedia.org/wiki/Torpedo_californica Torpedo californica]'' [[acetylcholinesterase]] (''Tc''AChE) complexed with [http://en.wikipedia.org/wiki/Tacrine tacrine] (THA), THA's [http://en.wikipedia.org/wiki/Acridine acridine] ring is stacked between the [http://en.wikipedia.org/wiki/Aromaticity aromatic rings] of <scene name='Demo_1acj/Oulu/2'>W84 and F330</scene>, near the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] of <scene name='1acj/Active_site_of_1acj/2'>AChE's active site</scene> which consists of '''S200''', '''E327''', '''H440'''. When comparing 3 recent complexes of ''Tc''AChE, i.e. edrophonium (EDR), decamethonium (DECA) and THA, the only major conformational difference between them is seen in the orientation of the [http://en.wikipedia.org/wiki/Phenyl_group phenyl ring] of F330. In the DECA complex it lies parallel to the surface of the gorge; in the other two complexes it is positioned to make contact with the bound ligand. This close interaction was confirmed by photoaffinity labeling by a 3H-labeled photosensitive probe, which labeled, predominantly, F330 within the active site. Labeling of <scene name='1acj/Tha_active_site_test_2/2'>W279</scene> was also observed. One mole of label is incorporated per mole of AChE inactivated, indicating that labeling of W279 and that of F330 are mutually exclusive. The structural and chemical data, together, show the important role of aromatic groups as binding sites for quaternary ligands, and they provide complementary evidence assigning W84 and F330 to the "anionic" subsite of the active site and W279 to the "peripheral" anionic site. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Huprine X==== | ||
| + | <scene name='1e66/Active_site/1'>Huprine X</scene> ('''HUP'''erzine A + tac'''RINE''') is one of the most potent [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor#Reversible_inhibitor reversible AChE inhibitors]. This <scene name='1e66/Active_site/2'>synthetic hybrid</scene> consists of a carbobicyclic moiety resembling that of (−)-<scene name='1e66/Active_site/12'>huperzine A</scene> <font color='blueviolet'><b>(colored blueviolet)</b></font> and the [http://en.wikipedia.org/wiki/4-Aminoquinoline 4-aminoquinoline] substructure of <scene name='1e66/Active_site/7'>tacrine</scene> <font color='magenta'><b>(colored magenta)</b></font>. Both these compounds are known AChE inhibitors. <font color='blueviolet'><b>(−)-Huperzine A</b></font> and <font color='magenta'><b>tacrine</b></font> positions partially overlap each other at the ''Tc''AChE <scene name='1e66/Active_site/13'>active site</scene>. ''Tc''AChE residues interacting with (−)-huperzine A ([[1vot]]) <span style="color:orange;background-color:black;font-weight:bold;">are colored orange</span> and with tacrine ([[1acj]]) <span style="color:cyan;background-color:black;font-weight:bold;">are colored cyan</span>. The <scene name='1e66/Active_site/10'>conformation</scene> of the 4-aminoquinoline substructure of the huprine X in its complex with ''Tc''AChE ([[1e66]], ''Tc''AChE interacting residues are in <span style="color:lime;background-color:black;font-weight:bold;">green</span>) is very similar to that of tacrine. The ring system of (−)-huperzine A is <scene name='1e66/Active_site/14'>rotated</scene> almost 180° relative to that of huprine X <ref name="Dvir">PMID:11863435</ref>. | ||
| + | {{Clear}} | ||
| + | |||
| + | ====Galanthamine==== | ||
| + | <scene name='AChE_inhibitors_and_substrates/Com_view_gal/1'>Galanthamine (GAL)</scene> [http://en.wikipedia.org/wiki/Galantamine]. <scene name='AChE_inhibitors_and_substrates/Com_view_gal/2'>GAL</scene> <font color='red'><b>(red)</b></font> is an [http://en.wikipedia.org/wiki/Alkaloid alkaloid] from the flower snowdrop ([http://en.wikipedia.org/wiki/Galanthus ''Galanthus nivalis'']). The [http://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystal structure] of the ''Tc''AChE/GAL complex ([[1dx6]]) was determined at 2.3 Å resolution. The inhibitor binds at the base of the [http://en.wikipedia.org/wiki/Active_site active site] gorge of ''Tc''AChE, interacting with both the choline-binding site (Trp84) and the acyl-binding pocket (Phe288, Phe290). The [http://en.wikipedia.org/wiki/Amine tertiary amine] appears to make a non-conventional [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond], via its N-methyl group, to Asp72. The [http://en.wikipedia.org/wiki/Hydroxyl#Hydroxyl_group hydroxyl group] of the inhibitor makes a strong hydrogen bond (2.7 Å) with Glu199 <ref name="Greenblatt">PMID:10606746</ref>. <font color='gray'><b>ACh (gray)</b></font> is shown for comparison. | ||
| + | |||
| + | {{Clear}} | ||
| + | ====Edrophonium==== | ||
| + | <scene name='2ack/Com_view/1'>Edrophonium (EDR)</scene> [http://en.wikipedia.org/wiki/Edrophonium] is stacked between the [http://en.wikipedia.org/wiki/Aromatic aromatic rings] of <scene name='2ack/Com_view/2'>W84 and F330</scene>, near the ''Tc''AChE <scene name='2ack/Com_view/3'>catalytic triad</scene> which consists of <font color='magenta'><b>'''S200'''</b></font>, <font color='magenta'><b>'''E327'''</b></font>, and <font color='magenta'><b>'''H440'''</b></font> ([[2ack]] or [[1ax9]]) <ref name="Ravelli">PMID:10089512</ref>. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:02, 8 March 2015
| |||||||||||
Treatment of Alzheimer's disease
Please see pages AChE inhibitors and substrates (Part II) and AChE inhibitors and substrates (Part III).
AChE bivalent inhibitors
Please see pages AChE bivalent inhibitors and AChE bivalent inhibitors (Part II)
Selected 3D Structures of AChE
- 2ace This is the original solved structure for Torpedo Californica
- 1ea5 This is one of the highest quality representative X-ray structures in the PDB.
- 1eve The E2020 (Aricept) complex.
- 1ax9 Endrophonium complex.
- 1vot Complex with Huperzine, a Chinese folk medicine.
- 1fss Complex with snake venum toxin Fasciculin-II.
- 1acj Complex with tacrine.
- 1e66 Complex with huprine X.
- 1dx6 Complex with galanthamine.
- 1qti Complex with galanthamine.
- 1w6r Complex with galanthamine iminium derivative.
- 2ack Complex with edrophonium.
- 1vzj Structure of the tetramerization domain of acetylcholinesterase.
- 1gqr Complex with rivastigmine.
- 1gqs Complex with NAP alone.
- 1vxr Complex with VX.
- 2vja Complex with OTMA.
- 1som Complex with soman.
- 2wfz Complex with nonaged soman.
- 2wg0 Complex with aged soman.
- 2wg1 Complex with aged soman and 2-PAM.
- 1cfj Complex with aged sarin.
- 2dfp Complex with aged DFP.
Additional Resources
For additional information, see: Alzheimer's Disease
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ 3.0 3.1 Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ 5.0 5.1 5.2 Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 1999 Jun 1;38(22):7032-9. PMID:10353814 doi:http://dx.doi.org/10.1021/bi982678l
- ↑ Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Dvir H, Wong DM, Harel M, Barril X, Orozco M, Luque FJ, Munoz-Torrero D, Camps P, Rosenberry TL, Silman I, Sussman JL. 3D structure of Torpedo californica acetylcholinesterase complexed with huprine X at 2.1 A resolution: kinetic and molecular dynamic correlates. Biochemistry. 2002 Mar 5;41(9):2970-81. PMID:11863435
- ↑ Greenblatt HM, Kryger G, Lewis T, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999 Dec 17;463(3):321-6. PMID:10606746
- ↑ Ravelli RB, Raves ML, Ren Z, Bourgeois D, Roth M, Kroon J, Silman I, Sussman JL. Static Laue diffraction studies on acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 1998 Nov 1;54(Pt 6 Pt 2):1359-66. PMID:10089512
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Michal Harel, Jaime Prilusky, David Canner