This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 996
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
<StructureSection load='1eci' size='340' side='right' caption='[[1eci]], [[NMR_Ensembles_of_Models | 20 NMR models]]' scene=''> | <StructureSection load='1eci' size='340' side='right' caption='[[1eci]], [[NMR_Ensembles_of_Models | 20 NMR models]]' scene=''> | ||
Ectatomin (1eci) is the main component of venom of the ant [https://en.wikipedia.org/wiki/Ectatomma_tuberculatum Ectatomma tuberculatum], making up 15%-18% of the crude venom and accounting for 90% of the venom's toxicity.<ref name="refthree" /> When bitten by E. tuberculatum, Ectatomin inserts into the target's [https://en.wikipedia.org/wiki/Cell_membrane cell membranes] and forms a nonselective [https://en.wikipedia.org/wiki/Ion_channel cation channel].<ref name="reftwo">PMID: 10336635</ref> The calculated [https://en.wikipedia.org/wiki/Isoelectric_point isoelectric point] and [https://en.wikipedia.org/wiki/Molecular_mass molecular weight] of Ectatomin are 9.95 and 7928 Da, respectively.<ref name="refthree">PMID: 8033986</ref> | Ectatomin (1eci) is the main component of venom of the ant [https://en.wikipedia.org/wiki/Ectatomma_tuberculatum Ectatomma tuberculatum], making up 15%-18% of the crude venom and accounting for 90% of the venom's toxicity.<ref name="refthree" /> When bitten by E. tuberculatum, Ectatomin inserts into the target's [https://en.wikipedia.org/wiki/Cell_membrane cell membranes] and forms a nonselective [https://en.wikipedia.org/wiki/Ion_channel cation channel].<ref name="reftwo">PMID: 10336635</ref> The calculated [https://en.wikipedia.org/wiki/Isoelectric_point isoelectric point] and [https://en.wikipedia.org/wiki/Molecular_mass molecular weight] of Ectatomin are 9.95 and 7928 Da, respectively.<ref name="refthree">PMID: 8033986</ref> | ||
| - | |||
== Structure == | == Structure == | ||
Biologically, Ectatomin exists as a heterodimer stabilized by <scene name='69/691538/Cysteine_disulfide/1'>disulfide linkages</scene> linkages. The <scene name='69/691538/Alpha_subunit/1'>α subunit</scene> has 37 amino acid residues, while the <scene name='69/691538/Beta_subunit/1'>β subunit</scene> has 34 amino acid residues. The structure of Ectatomin was solved using 2D NMR and CHARMm computational optimization, though there are 20 similar proposed models in total.<ref name="refone">PMID: 7881269</ref> | Biologically, Ectatomin exists as a heterodimer stabilized by <scene name='69/691538/Cysteine_disulfide/1'>disulfide linkages</scene> linkages. The <scene name='69/691538/Alpha_subunit/1'>α subunit</scene> has 37 amino acid residues, while the <scene name='69/691538/Beta_subunit/1'>β subunit</scene> has 34 amino acid residues. The structure of Ectatomin was solved using 2D NMR and CHARMm computational optimization, though there are 20 similar proposed models in total.<ref name="refone">PMID: 7881269</ref> | ||
| - | |||
Generally, each subunit is composed of two antiparallel α-helices, linked by disulfide bonds, with a connecting hairpin hinge region. The two subunits are linked by a disfulide bond between their hairpin hinge regions. One α-helix from each subunit is kinked approximately 40°, due to the presence of <scene name='69/691538/Prolines_both_subunits/2'>proline residues</scene>. The kinked α-helix of the α subunit is more kinked, containing three proline residues, while the kinked α-helix of the β subunit only contains one proline residue.<ref name="refone" /> | Generally, each subunit is composed of two antiparallel α-helices, linked by disulfide bonds, with a connecting hairpin hinge region. The two subunits are linked by a disfulide bond between their hairpin hinge regions. One α-helix from each subunit is kinked approximately 40°, due to the presence of <scene name='69/691538/Prolines_both_subunits/2'>proline residues</scene>. The kinked α-helix of the α subunit is more kinked, containing three proline residues, while the kinked α-helix of the β subunit only contains one proline residue.<ref name="refone" /> | ||
| - | |||
The internal region between the two subunits is primarily composed of hydrophobic residues. | The internal region between the two subunits is primarily composed of hydrophobic residues. | ||
| - | |||
Sequence - α subunit - GVIPKKIWETVCPTVEPWAKKCSGDIATYIKRECGKL<ref name="reftwo" /> | Sequence - α subunit - GVIPKKIWETVCPTVEPWAKKCSGDIATYIKRECGKL<ref name="reftwo" /> | ||
| Line 21: | Line 17: | ||
Sequence - β subunit - WSTIVKLTICPTLKSMAKKCEGSIATMIKKKCDK<ref name="reftwo" /> | Sequence - β subunit - WSTIVKLTICPTLKSMAKKCEGSIATMIKKKCDK<ref name="reftwo" /> | ||
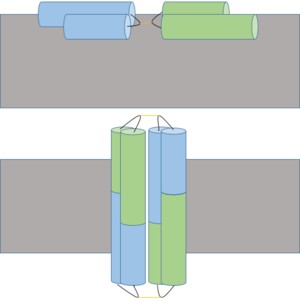
| + | == Mechanism == | ||
| - | + | [[Image:Possible_Ectatomin_Mechanism_3.png|300px|left|thumb| A proposed membrane insertion (above) and dimerization mechanism to form cation channel (below) of Ectatomin. Insertion occurs when the α and β subunits open at the hairpin hinge region (shown in black with yellow disulfide bonds), exposing internal hydrophobic residues which interact with hydrophobic lipid tails of the cell membrane. Pore formation occurs after dimerization, allowing ions to freely cross the membrane.]] | |
| - | + | ||
Ectatomin has several proposed mechanisms of action. The primary proposed mechanism involves the formation of a nonselective cation channel. In this mechanism, the α and β subunits open up, exposing the internal hydrophobic residues. The protein flattens while remaining attached at the hairpin hinge region. The now exposed hydrophobic residues nonselectively insert into plasma membranes. The inserted protein dimerizes, eventually forming a nonselective cation channel. | Ectatomin has several proposed mechanisms of action. The primary proposed mechanism involves the formation of a nonselective cation channel. In this mechanism, the α and β subunits open up, exposing the internal hydrophobic residues. The protein flattens while remaining attached at the hairpin hinge region. The now exposed hydrophobic residues nonselectively insert into plasma membranes. The inserted protein dimerizes, eventually forming a nonselective cation channel. | ||
| - | |||
For the second and third proposed mechanisms of action, Ectatomin has also been shown to inhibit kinases, specifically protein tyrosine kinase and protein kinase C, and Ca2+ channels. Kinase inhibition would potentially allow Ectatomin to interfere with various components of signal transduction. Calcium channel inhibition would potentially allow Ectatomin to affect physiological processes such as contraction, neurotransmitter release and neuronal activity regulation. | For the second and third proposed mechanisms of action, Ectatomin has also been shown to inhibit kinases, specifically protein tyrosine kinase and protein kinase C, and Ca2+ channels. Kinase inhibition would potentially allow Ectatomin to interfere with various components of signal transduction. Calcium channel inhibition would potentially allow Ectatomin to affect physiological processes such as contraction, neurotransmitter release and neuronal activity regulation. | ||
| - | |||
| - | [[Image:Possible_Ectatomin_Mechanism_3.png|300px|left|thumb| A proposed membrane insertion (above) and dimerization mechanism to form cation channel (below) of Ectatomin. Insertion occurs when the α and β subunits open at the hairpin hinge region (shown in black with yellow disulfide bonds), exposing internal hydrophobic residues which interact with hydrophobic lipid tails of the cell membrane. Pore formation occurs after dimerization, allowing ions to freely cross the membrane.]] | ||
== Toxicology == | == Toxicology == | ||
| Line 41: | Line 34: | ||
</StructureSection> | </StructureSection> | ||
| - | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 00:04, 11 March 2015
Ectatomin (1eci)
| |||||||||||
References
- ↑ 1.0 1.1 Arseniev AS, Pluzhnikov KA, Nolde DE, Sobol AG, Torgov MYu, Sukhanov SV, Grishin EV. Toxic principle of selva ant venom is a pore-forming protein transformer. FEBS Lett. 1994 Jun 27;347(2-3):112-6. PMID:8033986
- ↑ 2.0 2.1 2.2 Pluzhnikov K, Nosyreva E, Shevchenko L, Kokoz Y, Schmalz D, Hucho F, Grishin E. Analysis of ectatomin action on cell membranes. Eur J Biochem. 1999 Jun;262(2):501-6. PMID:10336635
- ↑ 3.0 3.1 3.2 3.3 Nolde DE, Sobol AG, Pluzhnikov KA, Grishin EV, Arseniev AS. Three-dimensional structure of ectatomin from Ectatomma tuberculatum ant venom. J Biomol NMR. 1995 Jan;5(1):1-13. PMID:7881269
- ↑ Touchard A, Labriere N, Roux O, Petitclerc F, Orivel J, Escoubas P, Koh JM, Nicholson GM, Dejean A. Venom toxicity and composition in three Pseudomyrmex ant species having different nesting modes. Toxicon. 2014 Sep;88:67-76. doi: 10.1016/j.toxicon.2014.05.022. Epub 2014 Jun 11. PMID:24929139 doi:http://dx.doi.org/10.1016/j.toxicon.2014.05.022