We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Amylase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
=Structure<ref name="Main"/>= | =Structure<ref name="Main"/>= | ||
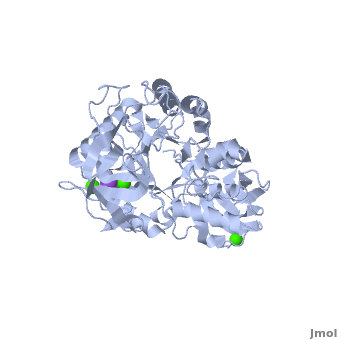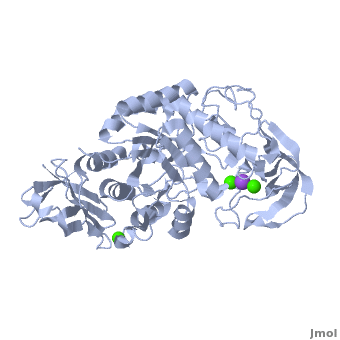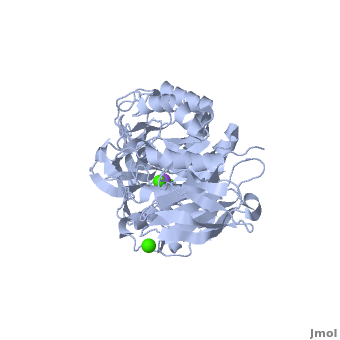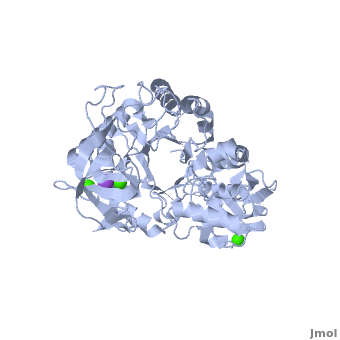
| - | Shown as 1hvx is the structure of the thermostable α-amylase of ''Bacillus stearothermophilus'' (BSTA)<ref name="Main"/>. BSTA is comprised of a single polypeptide chain. This chain is folded into three domains: A, B and C. These domains are generally found on all α-amylase enzymes. The <scene name='Sandbox_182/Domain_aa/1'>A domain </scene>constitutes the core structure, with a (β/α)<sub>8</sub>-barrel.The <scene name='Sandbox_182/Domain_a/1'> B domain</scene> consists of a sheet of four anti-parallel β-strands with a pair of anti-parallel β-strands. Long loops are observed between the β-strands. Located within the B domain is the <scene name='Sandbox_182/Trio/1'>binding site</scene> for Ca<sup>2+</sup>-Na<sup>+</sup>-Ca<sup>2+</sup>. <scene name='Sandbox_182/Domain_c/1'>Domain C </scene>consisting of eight β-strands is assembled into a globular unit forming a Greek key motif. It also holds the <scene name='Sandbox_182/Caiii/1'>third </scene>Ca<sup>2+</sup> binding site in association with domain A. Positioned on the C-terminal side of the β-strands of the (β/α)<sub>8</sub>-barrel in domain A is the active site. The catalytic residues involved for the BSTA active site are | + | Shown as 1hvx is the structure of the thermostable α-amylase of ''Bacillus stearothermophilus'' (BSTA)<ref name="Main"/>. BSTA is comprised of a single polypeptide chain. This chain is folded into three domains: A, B and C. These domains are generally found on all α-amylase enzymes. The <scene name='Sandbox_182/Domain_aa/1'>A domain </scene>constitutes the core structure, with a (β/α)<sub>8</sub>-barrel.The <scene name='Sandbox_182/Domain_a/1'> B domain</scene> consists of a sheet of four anti-parallel β-strands with a pair of anti-parallel β-strands. Long loops are observed between the β-strands. Located within the B domain is the <scene name='Sandbox_182/Trio/1'>binding site</scene> for Ca<sup>2+</sup>-Na<sup>+</sup>-Ca<sup>2+</sup>. <scene name='Sandbox_182/Domain_c/1'>Domain C </scene>consisting of eight β-strands is assembled into a globular unit forming a Greek key motif. It also holds the <scene name='Sandbox_182/Caiii/1'>third </scene>Ca<sup>2+</sup> binding site in association with domain A. Positioned on the C-terminal side of the β-strands of the (β/α)<sub>8</sub>-barrel in domain A is the active site. The catalytic residues involved for the BSTA active site are <scene name='38/382954/Active_site_ball_stick/2'>Asp234, Glu264, and Asp331</scene>. The residues are identical to other α-amylases, yet there are positional differences which reflect the flexible nature of catalytic resides. |
| - | <scene name=' | + | |
<scene name='Sandbox_182/Trio/1'>CaII and CaI with Na</scene> found in the interior of domain B and <scene name='Sandbox_182/Caiii/2'>CaIII </scene>at the interface of domain A and C, constitute the metal ion binding sites. All α-amylases contain one strongly conserved Ca<sup>2+</sup> ion for structural integrity and enzymatic activity.<ref name="chloride">PMID: 12021442</ref> CaI is consistent in α-amylases, however there are structural differences between the linear trio of CaI, CaII and Na in other enzymes. CaIII acts as a bridge between two loops, one from α6 of domain A, and one between β1 and β2 of domain C. | <scene name='Sandbox_182/Trio/1'>CaII and CaI with Na</scene> found in the interior of domain B and <scene name='Sandbox_182/Caiii/2'>CaIII </scene>at the interface of domain A and C, constitute the metal ion binding sites. All α-amylases contain one strongly conserved Ca<sup>2+</sup> ion for structural integrity and enzymatic activity.<ref name="chloride">PMID: 12021442</ref> CaI is consistent in α-amylases, however there are structural differences between the linear trio of CaI, CaII and Na in other enzymes. CaIII acts as a bridge between two loops, one from α6 of domain A, and one between β1 and β2 of domain C. | ||
==Chloride Dependent Enzymes== | ==Chloride Dependent Enzymes== | ||
Revision as of 01:50, 7 April 2015
| |||||||||||
3D structures of amylase
Updated on 07-April-2015
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press
- ↑ 2.0 2.1 Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998 Mar;7(3):564-72. PMID:9541387
- ↑ 3.0 3.1 3.2 Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability. J Biochem. 2001 Mar;129(3):461-8. PMID:11226887
- ↑ 4.0 4.1 4.2 Aghajari N, Feller G, Gerday C, Haser R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002 Jun;11(6):1435-41. PMID:12021442
- ↑ Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 6.0 6.1 Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 7.0 7.1 7.2 7.3 Kuriki T, Imanaka T. The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87(5):557-65. PMID:16232518
- ↑ 8.0 8.1 PPMID: 17713601
- ↑ Franco OL, Rigden DJ, Melo FR, Grossi-De-Sa MF. Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases. Eur J Biochem. 2002 Jan;269(2):397-412. PMID:11856298
- ↑ Yang RW, Shao ZX, Chen YY, Yin Z, Wang WJ. Lipase and pancreatic amylase activities in diagnosis of acute pancreatitis in patients with hyperamylasemia. Hepatobiliary Pancreat Dis Int. 2005 Nov;4(4):600-3. PMID:16286272
Proteopedia Page Contributors and Editors (what is this?)
Shane Riley, Michal Harel, Joel L. Sussman, Randi Woodbeck, Jaime Prilusky, Alexander Berchansky, Ann Taylor, Andrea Gorrell, David Canner