Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== '''Structure''' == | == '''Structure''' == | ||
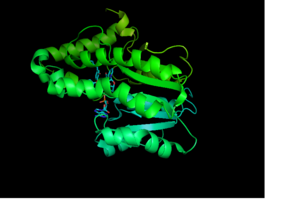
| - | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene>. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit features a single domain with a central core that supports a <scene name='69/694241/Monomer_subunit/2'>NADH binding site</scene>. | + | Crystal structures of InhA reveal a <scene name='69/694241/Homotetramer_subunits_labeled/1'>homotetramer</scene>. Each <scene name='69/694241/Monomer_subunit_no_ligands/1'>monomer</scene> subunit features a single domain with a central core that supports a <scene name='69/694241/Monomer_subunit/2'>NADH binding site</scene>. The alpha6 and alpha7 helices, composed of the <scene name='69/694241/Monomer_subunit_196_219/1'>amino acid residues 196-219</scene>, make up the substrate binding loop of InhA. |
| - | + | ||
| - | <scene name='69/694241/Monomer_subunit_196_219/1'>amino acid residues 196-219</scene> | + | |
<scene name='69/694241/Substrate_binding_pocket/1'>substrate binding pocket</scene> | <scene name='69/694241/Substrate_binding_pocket/1'>substrate binding pocket</scene> | ||
Revision as of 02:19, 7 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||