User:Braden Sciarra/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Structure == | == Structure == | ||
| - | The ICL homotetramer possesses | + | The ICL homotetramer possesses C2 symmetry, with an axis of rotation at x-axis, y-axis, and z-axis of the enzyme. Two individual subunits off ICL are held together by a characteristic <scene name='69/697526/Helix_swapping/3'>Helix Swapping</scene> between three alpha helices formed by residues 370-384, 349-367, and 399-409 on neighboring monomers. The interlocking mechanism created by these helices provides additional strength to hold the two monomeric subunits together, allowing ICL to essentially be composed of two dimerized subunits. This interaction will bury approximately 18% of the surface of each subunit, and will help to shield the interior binding site from hydration. |
| - | + | [[Image:homotetramer.png|150 px|right|thumb|Figure 2: C2 Symmetry of the homotetramer isocitrate lyase]] | |
| - | + | ||
| - | [[Image:homotetramer.png|150 px|right|thumb|C2 Symmetry]] | + | |
== Active Site == | == Active Site == | ||
| + | |||
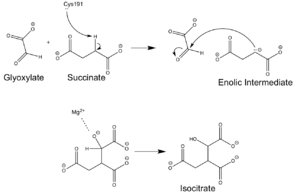
| + | The catalytic site of isocitrate lyase lies near the C-terminal ends of the Beta-strands of the active site. The glyoxylate substrate is held into place by a network of hydrogen bonds with Ser 91, Gly 92, Trp 93, and Arg 228. The magnesium ion serves to stabilize the partial negative charge placed on the carbonyl oxygens of the glyoxylate. The other substrate, succinate, contains two carboxyl groups and possesses a similar network of hydrogen bonding that holds the ligand in place within the active site. One carboxylate group is hydrogen bound to Asn 313, Glu 295, Arg 228, and Gly 192. The second carboxylate is hydrogen bound to Thr 347, Asn 313, Ser 315, Ser 317, and His 193. The ordered hydrogen bonding within the active site orients the succinate molecule such that the carbonyl carbon is only 3.2 angstroms away from the nucleophilic thiol group of Cys 191. | ||
<scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> | <scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> | ||
Revision as of 22:50, 7 April 2015
Isocitrate Lyase from Mycobacterium Tuberculosis
| |||||||||||