User:Braden Sciarra/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
===Catalytic Loop=== | ===Catalytic Loop=== | ||
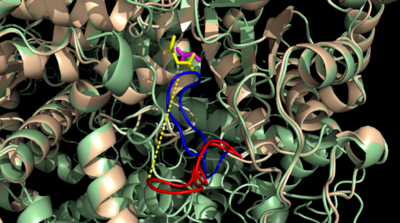
[[Image:Openvsclosed..png|400 px|left|thumb|Figure 3: Highly ordered hydrogen bonding network within the active site of ICL]] | [[Image:Openvsclosed..png|400 px|left|thumb|Figure 3: Highly ordered hydrogen bonding network within the active site of ICL]] | ||
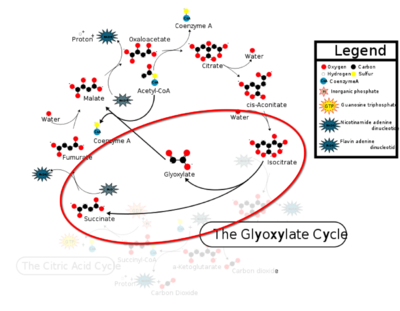
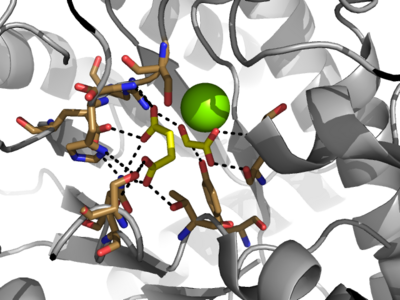
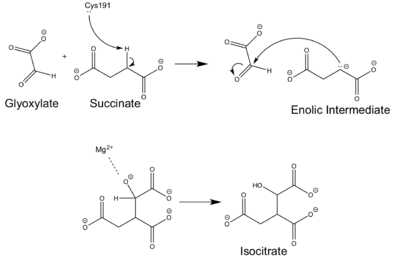
| - | The <scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> of the Isocitrate Lyase enzyme is composed of residues 185-196, and can exist in both the open and closed conformation. In the open conformation, the catalytic loop is oriented such that the catalytic CYS191 residue is located far from the active site, allowing for solvent accessibility and substrate binding.<ref name="solvent">Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 DOI: 10.1126/science.6879170</ref> Upon substrate binding, the catalytic loop adopts a closed conformation, moving between ten and fifteen angstroms<ref name="ICL">PMID:10932251</ref>. This closed conformation will cause the binding site to become inaccessible to the solvent. The loop closure is triggered by the movement of the Mg2+ ion that occurs upon binding of the succinate. This movement of the Mg2+ ion results in electrostatic interactions at LYS189, causing the loop to close. | + | The <scene name='69/697526/Catalytic_loop/1'>Catalytic Loop</scene> of the Isocitrate Lyase enzyme is composed of residues 185-196, and can exist in both the open and closed conformation. In the open conformation, the catalytic loop is oriented such that the catalytic CYS191 residue is located far from the active site, allowing for solvent accessibility and substrate binding.<ref name="solvent">Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 (1983). DOI: 10.1126/science.6879170</ref> Upon substrate binding, the catalytic loop adopts a closed conformation, moving between ten and fifteen angstroms<ref name="ICL">PMID:10932251</ref>. This closed conformation will cause the binding site to become inaccessible to the solvent. The loop closure is triggered by the movement of the Mg2+ ion that occurs upon binding of the succinate. This movement of the Mg2+ ion results in electrostatic interactions at LYS189, causing the loop to close. |
==Mechanism== | ==Mechanism== | ||
Revision as of 00:43, 8 April 2015
Isocitrate Lyase from Mycobacterium Tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr, Sacchettini JC. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000 Aug;7(8):663-8. PMID:10932251 doi:10.1038/77964
- ↑ 2.0 2.1 2.2 Beeching JR. High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis. Protein Seq Data Anal. 1989 Dec;2(6):463-6. PMID:2696959
- ↑ 3.0 3.1 3.2 3.3 Masamune et al. Bio-Claisen condensation catalyzed by thiolase from Zoogloea ramigera. Active site cysteine residues. "Journal of the American Chemical Society" 111: 1879-1881 (1989). DOI: 10.1021/ja00187a053
- ↑ Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 (1983). DOI: 10.1126/science.6879170