Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
Talk generally about catalytic triad. More specific details in sections below. | Talk generally about catalytic triad. More specific details in sections below. | ||
| + | ==='''Ligands'''=== | ||
| + | |||
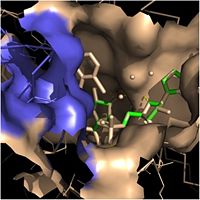
| + | The two ligands that play a role in the function of InhA are [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide NADH] and the long-chain fatty acyl substrate. NADH, or nicotinamide adenine dinucleotide, is a cofactor found in all living cells. It is especially important in the function of InhA because it helps position the fatty acyl substrate within the fatty acyl binding crevice, thereby improving the overall activity of InhA. Once properly positioned, the fatty acyl substrates are reduced at the ''trans'' double bond between C2 and C3, forming mycolic acid precursors that eventually compose the cell wall of mycobacteria. | ||
=== '''Fatty Acyl Binding Crevice''' === | === '''Fatty Acyl Binding Crevice''' === | ||
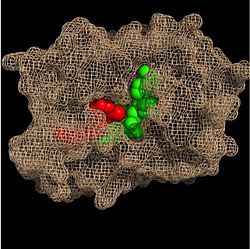
| - | Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Both of these substrates are held in place through interactions with the side chains of <scene name='69/694241/Sbl_hydrophobic/1'>hydrophobic</scene> (purple) residues. [[Image:Binding Pocket - Mesh.jpg|thumb|250px|right|Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)]] The majority of these <scene name='69/694241/Hydrophobic_residues/1'>hydrophobic</scene> residues anchoring the substrates are found within the substrate binding loop itself, including Ala-198, Met-199, Ala-201, Ile-202, Leu-207, Ile-215, and Leu-218. Studies have found that the fatty acyl substrate adopts a u-shaped conformation to facilitate binding. | + | Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Both of these substrates are held in place through interactions with the side chains of <scene name='69/694241/Sbl_hydrophobic/1'>hydrophobic</scene> (purple) residues. [[Image:Binding Pocket - Mesh.jpg|thumb|250px|right|Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)]] The majority of these <scene name='69/694241/Hydrophobic_residues/1'>hydrophobic</scene> residues anchoring the substrates are found within the substrate binding loop itself, including Ala-198, Met-199, Ala-201, Ile-202, Leu-207, Ile-215, and Leu-218. Studies have found that the fatty acyl substrate adopts a u-shaped conformation to facilitate binding. Additional hydrophobic amino acids that are not a part of the substrate binding loop also play a role in positioning and stabilizing the fatty acyl chain in the crevice. |
Revision as of 01:38, 8 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||