Sandbox Reserved 1074
From Proteopedia
(Difference between revisions)
| Line 35: | Line 35: | ||
==='''Substrate Binding Loop Flexibility''' === | ==='''Substrate Binding Loop Flexibility''' === | ||
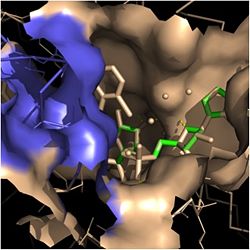
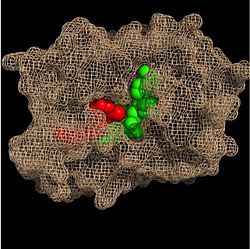
| - | In addition to the hydrogen bonding that occurs between the NADH and fatty acyl substrate within the crevice, each of these molecules is also held in place within the crevice through interactions with the side chains of surrounding <scene name='69/694241/Sbl_hydrophobic/1'>hydrophobic</scene> (purple) residues. The majority of these <scene name='69/694241/Hydrophobic_residues/1'>hydrophobic</scene> residues anchoring the substrates are found within the substrate binding loop itself, including Ala-198, Met-199, Ala-201, Ile-202, Leu-207, Ile-215, and Leu-218. Additional hydrophobic amino acids that are not a part of the substrate binding loop also play a role in positioning and stabilizing the fatty acyl chain in the crevice. Studies have found that the fatty acyl substrate adopts a u-shaped conformation to facilitate binding. As the fatty acyl substrate binds in the crevice, the substrate binding loop shifts outward toward the solvent, and the [[ | + | In addition to the hydrogen bonding that occurs between the NADH and fatty acyl substrate within the crevice, each of these molecules is also held in place within the crevice through interactions with the side chains of surrounding <scene name='69/694241/Sbl_hydrophobic/1'>hydrophobic</scene> (purple) residues. The majority of these <scene name='69/694241/Hydrophobic_residues/1'>hydrophobic</scene> residues anchoring the substrates are found within the substrate binding loop itself, including Ala-198, Met-199, Ala-201, Ile-202, Leu-207, Ile-215, and Leu-218. Additional hydrophobic amino acids that are not a part of the substrate binding loop also play a role in positioning and stabilizing the fatty acyl chain in the crevice. Studies have found that the fatty acyl substrate adopts a u-shaped conformation to facilitate binding. As the fatty acyl substrate binds in the crevice, the substrate binding loop shifts outward toward the solvent, and the [[#Importance of Tyr-158|Tyr-158]] residue is rotated to facilitate the binding of fatty acyl chains of 16 carbons or greater. During this conformational change upon substrate binding, no hydrogen bonds are broken, which supports the flexibility of the substrate binding loop. It is likely that this flexibility of the substrate binding loop provides increased freedom for the active site to accept fatty acyl substrates of varying carbon chain lengths <ref name="InhA" />. |
==='''Importance of Tyr-158'''=== | ==='''Importance of Tyr-158'''=== | ||
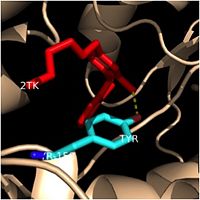
| - | One example of a hydrophobic amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is | + | One example of a hydrophobic amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is Tyr-158. [[Image:Tyr-158.jpg|thumb|200px|left|Figure 4. Tyr-158 (light blue) hydrogen bonding to the fatty acyl substrate, 2TK (red)]] This amino acid is conserved in other enoyl-ACP reductases in both bacteria and plants, so it likely plays an essential role in the function of these specific enzymes. Studies have shown that Tyr-158 forms the only direct hydrogen bond that exists between the InhA protein and the fatty acyl substrate. This hydrogen bond occurs between the hydroxyl oxygen on the side chain of Tyr-158 and the thioester carbonyl oxygen of the fatty acyl substrate. Consequently, the Tyr-158 residue acts to stabilize the enolate intermediate that forms during the hydride transfer reaction <ref name="InhA" />. |
| Line 51: | Line 51: | ||
=== Isoniazid === | === Isoniazid === | ||
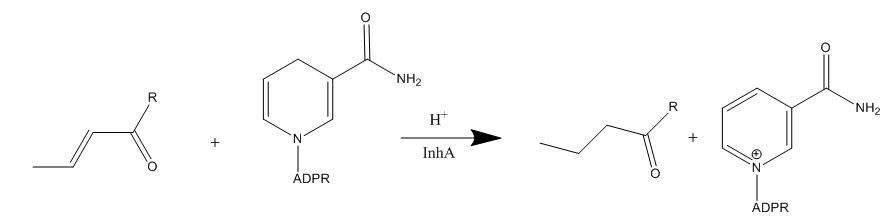
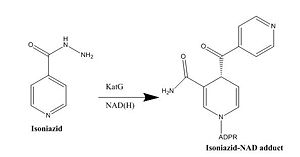
| - | Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser | + | Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser-94 to Ala of InhA was sufficient enough to have isoniazid resistance <ref name="InhA" />. |
=== Other Inhibitors === | === Other Inhibitors === | ||
| - | Drug resistance of ''M. tuberculosis'' has become a huge problem for the development of antibiotics. A drug screen of potential inhibitors of InhA (300 compounds) was composed of inhibitors of the [http://en.wikipedia.org/wiki/Plasmodium_falciparum ''Plasmodium falciparum''] enoyl-reductase. The enoyl reductases of both bacteria have limited similarities, however two compounds, CD39 and CD117 had activity against drug-susceptible ''M. tuberculosis''. More importantly, both compounds had activity against drug-resistant and multi-drug resistant TB. Treatment of the bacterium with the compounds resulted in the inhibition of mycolic acid and long-chain fatty acid biosynthesis, indicating that these compounds act against enzymes of both the FAS-I and FAS-II system. The benefit of having the compounds have multiple targets is the reduced development of drug resistance, which is the disadvantage of isoniazid. The essential chemical groups that lead to the antimycobacterial properties of the compounds include a [http://en.wikipedia.org/wiki/Thioacetic_acid thioacetate] group, and a [http://en.wikipedia.org/wiki/Butyl t-butyl] group <ref name="other inhibitors"> Vilchèze, C. ''et al.'' (2011). Novel Inhibitors of InhA Efficiently Kill Mycobacterium tuberculosis under Aerobic and Anaerobic Conditions. Antimicrobial Agents and Chemotherapy, 55(8),''3889-3898. DOI: [http://aac.asm.org/content/55/8/3889.full#T1 10.1128/AAC.00266-11]</ref>. | + | Drug resistance of ''M. tuberculosis'' has become a huge problem for the development of antibiotics. A drug screen of potential inhibitors of InhA (300 compounds) was composed of inhibitors of the [http://en.wikipedia.org/wiki/Plasmodium_falciparum ''Plasmodium falciparum''] enoyl-reductase. The enoyl reductases of both bacteria have limited similarities, however two compounds, CD39 and CD117 had activity against drug-susceptible ''M. tuberculosis''. More importantly, both compounds had activity against drug-resistant and multi-drug resistant TB. Treatment of the bacterium with the compounds resulted in the inhibition of mycolic acid and long-chain fatty acid biosynthesis, indicating that these compounds act against enzymes of both the FAS-I and FAS-II system. The benefit of having the compounds have multiple targets is the reduced development of drug resistance, which is the disadvantage of isoniazid. The essential chemical groups that lead to the antimycobacterial properties of the compounds include a [http://en.wikipedia.org/wiki/Thioacetic_acid thioacetate] group, and a [http://en.wikipedia.org/wiki/Butyl t-butyl] group <ref name="other inhibitors"> Vilchèze, C. ''et al.'' (2011). Novel Inhibitors of InhA Efficiently Kill Mycobacterium tuberculosis under Aerobic and Anaerobic Conditions. ''Antimicrobial Agents and Chemotherapy, 55(8),'' 3889-3898. DOI: [http://aac.asm.org/content/55/8/3889.full#T1 10.1128/AAC.00266-11]</ref>. |
[[Image:CD39.JPG|thumb|500px|left|Figure 6. CD39 Structure <ref name="other inhibitors">]] [[Image:CD117.JPG|thumb|600 px|center|Figure 7. CD117 Structure <ref name="other inhibitors">]] | [[Image:CD39.JPG|thumb|500px|left|Figure 6. CD39 Structure <ref name="other inhibitors">]] [[Image:CD117.JPG|thumb|600 px|center|Figure 7. CD117 Structure <ref name="other inhibitors">]] | ||
Revision as of 23:30, 8 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Bell, A.F. et al.(2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. Journal of the American Chemical Society, 129, 6425-6431. DOI: 10.1021/ja068219m
- ↑ 2.0 2.1 Bhatt, A. et al. (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Journal of Molecular Microbiology, 64(6), 1442-1454. PMID: 17555433 DOI: 10.1111/j.1365-2958.2007.05761.x
- ↑ Marrakchi, Hedia, et al. (2000). InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Journal of Microbiology, 146, 289-296. PMID: 10708367
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Rozwarski, D.A. et al. (1999). Crystal Structure of the Mycobacterium tuberculosis Enoyl-ACP Reductase, InhA, in Complex with NAD+ and a C16 Fatty Acyl Substrate. Journal of Biological Chemistry, 274(22), 15582-15589. PMID: 10336454 DOI: 10.1074/jbc.274.22.15582
- ↑ 5.0 5.1 Vilchèze, C. et al. (2011). Novel Inhibitors of InhA Efficiently Kill Mycobacterium tuberculosis under Aerobic and Anaerobic Conditions. Antimicrobial Agents and Chemotherapy, 55(8), 3889-3898. DOI: 10.1128/AAC.00266-11