Sandbox Reserved 1066
From Proteopedia
| Line 14: | Line 14: | ||
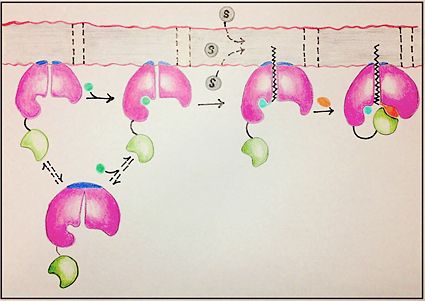
[[Image:FadD13 edited image.jpg|425 px|left|thumb|Figure 1: Mechanism for the activation of fatty acids (C24-C26) by FadD13. The N terminal domain (pink) is embedded in the membrane with the arginine rich lid-loop (dark blue), while the flexile linker (black) connects the C terminal domain (green) to the rest of the enzyme. Activation requires the binding of ATP (blue) which induces structural changes that promote the binding of the fatty acid chain. Formation of an acyl-adenylate intermediate induces a 140° rotation of the C terminal domain and the binding of CoA (orange). ]] | [[Image:FadD13 edited image.jpg|425 px|left|thumb|Figure 1: Mechanism for the activation of fatty acids (C24-C26) by FadD13. The N terminal domain (pink) is embedded in the membrane with the arginine rich lid-loop (dark blue), while the flexile linker (black) connects the C terminal domain (green) to the rest of the enzyme. Activation requires the binding of ATP (blue) which induces structural changes that promote the binding of the fatty acid chain. Formation of an acyl-adenylate intermediate induces a 140° rotation of the C terminal domain and the binding of CoA (orange). ]] | ||
| - | <scene name='69/694233/Lys_487/2'>Lys 487</scene> results in a 95% loss of function of FadD13. <ref name="residue paper">PMCID: | + | <scene name='69/694233/Lys_487/2'>Lys 487</scene> results in a 95% loss of function of FadD13. <ref name="residue paper">PMCID: PMC2793005ref> |
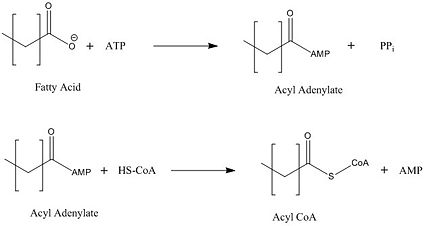
[[Image:acyl coa synthetase.jpg|425 px|left|thumb|Figure 2: Representation of the two-step reaction catalyzed by FadD13]] | [[Image:acyl coa synthetase.jpg|425 px|left|thumb|Figure 2: Representation of the two-step reaction catalyzed by FadD13]] | ||
Revision as of 00:58, 9 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase
| |||||||||||
References
- ↑ Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Molina DM, Hogbom M. The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme. Structure. 2012 May 2. PMID:22560731 doi:10.1016/j.str.2012.03.012
- ↑ Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis--a potential target for the development of antitubercular drugs. J Mol Model. 2011 Feb;17(2):301-13. doi: 10.1007/s00894-010-0727-3. Epub 2010 May, 8. PMID:20454815 doi:http://dx.doi.org/10.1007/s00894-010-0727-3
- ↑ PMCID: PMC2793005ref>
Contents
General mechanism for the activation of fatty acids
FadD13 first activates the fatty acid through a reaction with ATP to form an acyl adenylate intermediate and release pyrophosphate. Following a conformational change of the enzyme, coenzyme A is able to bind and reaction with the acyl adenylate intermediate forming the acyl CoA product (Figure 2).
Structural basis for housing lipid substrates longer than the enzyme
The ability for FadD13 to transport and activate fatty acids of the maximum tested length C26, lies in it being a peripheral membrane protein with a hydrophobic tunnel. FadD13 is attached to the membrane via electrostatic interactions in the N-terminal domain. Of importance in the region is the arginine rich lid-loop which serves to block the transport of fatty acids into the enzyme. Once the lid-loop is opened, fatty acids may be pulled from the membrane into a hydrophobic tunnel, which is the main structural component by which fatty acids are transported from the membrane into the cell. Negatively charged residues at the active site of FadD13 are the driving factor in the attraction of the fatty acid from the membrane through the hydrophobic tunnel of the enzyme.
Structure
FadD13 is composed of 503 amino acid residues divided into three main regions: The (residues 1-395) and (residues 402-503) which are connected via a flexible represented in dark blue (residues 396-401).<ref></ref> Each region plays an important role in the activation of fatty acids. The large N-terminal domain has the most structural features, but it is ultimately the flexible linker that allows movement of the C-terminal domain to from the fully functioning active site (Figure 1).
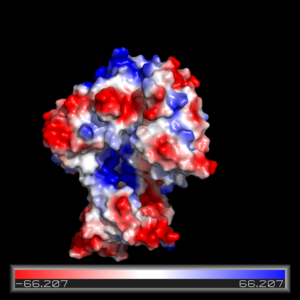
Electrostatics
The electrostatics of FadD13 as seen in (Fiigure 3) illustrate the hydrophobic and positively charged regions that compose this protein. Experimental results revealed that the peripheral FadD13 is attached to the membrane via electrostatic and hydrophobic regions located on the top portion of the N-terminal region (figure 3).<ref></ref> Of key importance in this N-terminal domain region attached to the membrane is an area of notable arginine rich residues, known as the arginine rich lid-loop.
Arginine Rich Lid-loop
The functions to block entry of fatty acids into the hydrophobic tunnel of FadD13.This area on the top portion of the enzyme is also crucial in the association with the membrane as the positively charged arginine residues are attracted to the negative charge on the phospholipid heads.<ref></ref>
Hydrophobic Tunnel
The hydrophobic tunnel of FadD13 is essential to the transport and accommodation of very long fatty acids from the membrane into the cell. This tunnel runs through the middle of FadD13 from the arginine rich lid loop to the ATP binding site and is situated between the and alpha helices α8-α9 and parallel beta sheet β9- β14.<ref></ref>
Active Site
The active site on FadD13 is composed of two conserved regions, one of which serves as the binding site for ATP and the other for CoA. The adenine of ATP is bound to a group of that is structurally identically to other acyl-CoA synthetases. <ref></ref>
Disease
Mycobacterium tuberculosis is the causative agent involved in the disease tuberculosis.
Relevance
Inhibitors
Future Research
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
Similar Proteopedia Pages
Thioesterase Do we have anything like this?
External Resources
Tuberculosis Wikipedia page
Mycobacterium tuberculosis Wikipedia page
Coenzyme A Wikipedia page
Acyl CoA Wikipedia Page
Mycolic Acid Wikipedia page