We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1071
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
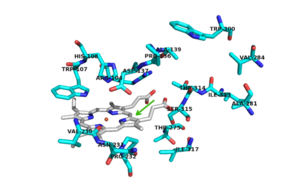
There are 6 conserved key active site residues that suround the <scene name='69/694238/Heme/2'>heme</scene>. These <scene name='69/694238/Active_site/2'>active site</scene> residues are Arg 104, Trp 107, His 108, His 270, Asp 381. | There are 6 conserved key active site residues that suround the <scene name='69/694238/Heme/2'>heme</scene>. These <scene name='69/694238/Active_site/2'>active site</scene> residues are Arg 104, Trp 107, His 108, His 270, Asp 381. | ||
| + | |||
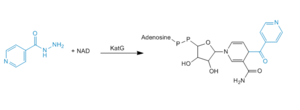
| + | [[Image:INH_mechanism.PNG|300 px|left|thumb|Figure Legend]] | ||
The location of the binding site for [http://en.wikipedia.org/wiki/Isoniazid isoniazid (INH)] is located near the ''δ meso'' heme edge, about 3.8 Å away from the heme iron. This binding site is found within what is considered to be the usual substrate access channel of peroxidases. The reaction between INH and the enzyme must occur from interaction in a binding site intended for the natural substrate (A2). Asp 137 plays a key role in the activation and binding of INH. Asp 137 creates energetically favorable interactions due to its ability to make hydrogen-bond interactions between its carboxylic acid side chain and the pyridinyl N1 of INH. | The location of the binding site for [http://en.wikipedia.org/wiki/Isoniazid isoniazid (INH)] is located near the ''δ meso'' heme edge, about 3.8 Å away from the heme iron. This binding site is found within what is considered to be the usual substrate access channel of peroxidases. The reaction between INH and the enzyme must occur from interaction in a binding site intended for the natural substrate (A2). Asp 137 plays a key role in the activation and binding of INH. Asp 137 creates energetically favorable interactions due to its ability to make hydrogen-bond interactions between its carboxylic acid side chain and the pyridinyl N1 of INH. | ||
| Line 36: | Line 38: | ||
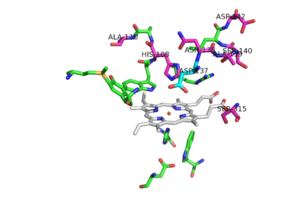
No known mutants have been reported to occur at Asp 137, although a few mutants nearby could cause local conformational changes and thereby altering the orientation of the Asp 137 side chain, making it less effective in binding and activation of INH. | No known mutants have been reported to occur at Asp 137, although a few mutants nearby could cause local conformational changes and thereby altering the orientation of the Asp 137 side chain, making it less effective in binding and activation of INH. | ||
| + | |||
| + | [[Image:Mutation_locations.png|300 px|left|thumb|Pink residues represent the location of possible mutations. Green residues represent the active site. Asp 137 is shown in blue.]] | ||
Revision as of 01:45, 9 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Mycobacterium tuberculosis Catalase-Peroxidase
| |||||||||||