We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1061
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
The active site of the protein is stabilized through a network of hydrogen bonds involving the two highly conserved residues, CVQC and WSGFRP. The crystal structure shows that interactions with one water molecule is necessary for the proper coordination between the conserved motifs to occur. These hydrogen bonds orient the important residues in the most optimal position to promote oxidation and reduction. | The active site of the protein is stabilized through a network of hydrogen bonds involving the two highly conserved residues, CVQC and WSGFRP. The crystal structure shows that interactions with one water molecule is necessary for the proper coordination between the conserved motifs to occur. These hydrogen bonds orient the important residues in the most optimal position to promote oxidation and reduction. | ||
| + | |||
| + | [[Image:Weblogocvqc.png|thumb|center|upright=2.5|Weblogo diagram showing highly conserved CVQC region of NrdH.]] | ||
<scene name='69/694227/Arg_68_conformation_2/1'>TextToBeDisplayed</scene> | <scene name='69/694227/Arg_68_conformation_2/1'>TextToBeDisplayed</scene> | ||
Revision as of 00:23, 10 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
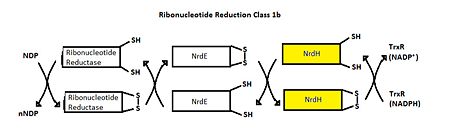
Structure of Mycobacterium Tuberculosis NrdH
| |||||||||||
References
- ↑ Makhlynets, O., Boal, A. K., Rhodes, D. V., Kitten, T., Rosenzweig, A. C., & Stubbe, J. (2014). Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS. The Journal of Biological Chemistry, 289(9), 6259–6272. doi:10.1074/jbc.M113.533554.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644