Sandbox Reserved 1058
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
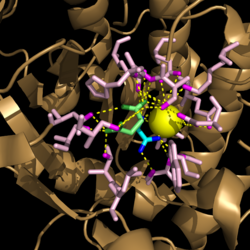
[[Image:Active_Site_Hydrogen_Bonding.png|250 px|left|thumb|'''Figure 3. Active site residues hydrogen bound to a cofactor and the products of the catalyzed isocitrate reaction.''' Glyoxylate is shown in blue, succinate is shown in green, and the Mg<sup>2+</sup> cofactor is shown in yellow.]] | [[Image:Active_Site_Hydrogen_Bonding.png|250 px|left|thumb|'''Figure 3. Active site residues hydrogen bound to a cofactor and the products of the catalyzed isocitrate reaction.''' Glyoxylate is shown in blue, succinate is shown in green, and the Mg<sup>2+</sup> cofactor is shown in yellow.]] | ||
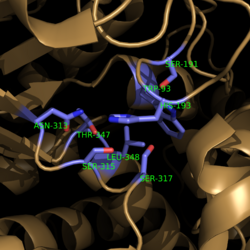
| - | [[Image:Active Site Residues.png|250 px| | + | [[Image:Active Site Residues.png|250 px|center|thumb|'''Figure 4. Active Site Residues.''' All eight active site residues necessary for catalysis of isocitrate are shown in slate. However, the protein shown is a C191S mutant of isocitrate lyase.]] |
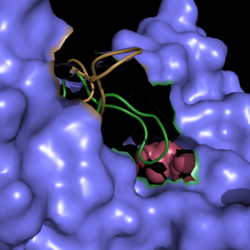
[[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 2. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop unbound is shown in wheat and the active site loop bound is shown in green.]] | [[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 2. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop unbound is shown in wheat and the active site loop bound is shown in green.]] | ||
| Line 25: | Line 25: | ||
==Mechanism of Action== | ==Mechanism of Action== | ||
| - | [[Image:TCA_Cycle.png|500 px| | + | [[Image:TCA_Cycle.png|500 px|left|thumb|'''Figure 5. Citric Acid Cycle with Glyoxylate Shunt Pathway.''' In several bacterial species, there is a carbon conserving gloxylate shunt pathway that converts isocitrate to malate in two steps instead of the usual five steps.]] |
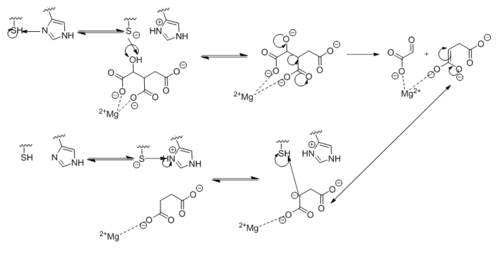
| - | [[Image:Complete_Mechanism.PNG|500 px| | + | [[Image:Complete_Mechanism.PNG|500 px|left|thumb|'''Figure 6. Observed Mechanism for the Breakdown of Isocitrate by Isocitrate Lyase.''' His193 shifts the pKa of Cys191 and removes its proton. This allows Cys191 to extract a proton from the hydroxyl group of isocitrate. The resulting oxyanion forms a carbonyl and forces the lysis of a C-C bond. Glyoxylate and the enol form of succinate are formed and stabilized with a Mg<sup>2+</sup> ion. The succinate enolate resonates and extracts the proton back from Cys191 to form succinate.]] |
| + | |||
Revision as of 03:01, 10 April 2015
Isocitrate Lyase from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annual Review of Microbiology. 1998, 52:127-164. doi: 10.1146/annurev.micro.52.1.127.
- ↑ Srivastava, V.; Janin, A.; Srivastava, B.; Srivastava, R.; Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. ScienceDirect. 2007. doi:10.1016/j.tube.2007.10.002.
- ↑ Muñoz-Elías, E.; McKinney, J.; M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005. 11(6):638-644. doi:10.1038/nm1252.
- ↑ Dunn, M.; Ramírez-Trujillo, J.; Hernández-Lucas, I.; Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009. 155:3166-3175. doi:10.1099/mic.0.030858-0.