We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1058
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
Isocitrate lyase is a [[lyase]] found in the proteome of multiple bacteria that oxidizes the hydroxl group of [https://en.wikipedia.org/wiki/Isocitric_acid isocitrate] and cleaves the substrate in two forming [https://en.wikipedia.org/wiki/Glyoxylic_acid glyoxylate] and [https://en.wikipedia.org/wiki/Succinic_acid succinate]. Isocitrate lyase is a tetramer that is composed primarily of alpha helices and beta sheets with a unique structural phenomenon called "<scene name='69/694225/Helix_swapping/1'>helix swapping</scene>". This enzyme can be found within the cytosol of bacteria and is used in a variation of the citric acid cycle to help conserve energy by not using [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide_phosphate NADPH] as an electron carrier and by reforming [http://en.wikipedia.org/wiki/Coenzyme_A coenzyme-A] earlier than in the normal citric acid cycle. | Isocitrate lyase is a [[lyase]] found in the proteome of multiple bacteria that oxidizes the hydroxl group of [https://en.wikipedia.org/wiki/Isocitric_acid isocitrate] and cleaves the substrate in two forming [https://en.wikipedia.org/wiki/Glyoxylic_acid glyoxylate] and [https://en.wikipedia.org/wiki/Succinic_acid succinate]. Isocitrate lyase is a tetramer that is composed primarily of alpha helices and beta sheets with a unique structural phenomenon called "<scene name='69/694225/Helix_swapping/1'>helix swapping</scene>". This enzyme can be found within the cytosol of bacteria and is used in a variation of the citric acid cycle to help conserve energy by not using [http://en.wikipedia.org/wiki/Nicotinamide_adenine_dinucleotide_phosphate NADPH] as an electron carrier and by reforming [http://en.wikipedia.org/wiki/Coenzyme_A coenzyme-A] earlier than in the normal citric acid cycle. | ||
==Isocitrate Lyase== | ==Isocitrate Lyase== | ||
| - | === | + | ===Structure=== |
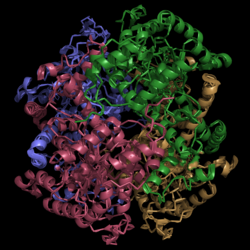
[[Image:Normal_Crystal_Structure.png|250 px|left|thumb|'''Figure 1. Crystal Structure of Isocitrate Lyase.''' Quaternary structure is comprised of four subunits forming an alpha/beta barrel.]] | [[Image:Normal_Crystal_Structure.png|250 px|left|thumb|'''Figure 1. Crystal Structure of Isocitrate Lyase.''' Quaternary structure is comprised of four subunits forming an alpha/beta barrel.]] | ||
[http://www.rcsb.org/pdb/explore/explore.do?structureId=1f8i Isocitrate lyase] is a tetramer with 222 symmetry. Each subunit is composed of 14 alpha helices and 14 beta sheets which includes a total of 426 residues. These α helices and β sheets form an unusual α/β barrel seen in Figure 1. The α/β barrel contains a topology of (βα)<sub>2</sub>α(βα)<sub>5</sub>β, differing from the canonical (βα)<sub>8</sub> pattern. Residues 184-200 and 235-254 connects the third and forth β-strands to their consecutive helices and form a <scene name='69/694225/Small_beta_domain/1'>small β-domain</scene> that consists of a short five-stranded βsheet (β6,β7,β9,β10,β11) that lies on top of the α/β barrel. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> A study of the equilibria between the four subunits shows that each isocitrate lyase monomer has a dynamic comformational change of the active site loop. At any given time, only two of the subunits are in the open conformation. <ref name="gould"> Gould, T.; van de Langemheen, H.; Muñoz-Elías, E.; McKinney, D.; Sacchettini, J.; Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in ''Mycobacterium tuberculosis''. ''Molecular Microbiology''. '''2006'''. ''61(4)'':940-947. doi:10.1111/j.1365-2958.2006.05297.x. </ref> Isocitrate Lyase shows a resemblance to [http://www.rcsb.org/pdb/explore/explore.do?structureId=1S2V phosphoenolpyrvate mutase]. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> | [http://www.rcsb.org/pdb/explore/explore.do?structureId=1f8i Isocitrate lyase] is a tetramer with 222 symmetry. Each subunit is composed of 14 alpha helices and 14 beta sheets which includes a total of 426 residues. These α helices and β sheets form an unusual α/β barrel seen in Figure 1. The α/β barrel contains a topology of (βα)<sub>2</sub>α(βα)<sub>5</sub>β, differing from the canonical (βα)<sub>8</sub> pattern. Residues 184-200 and 235-254 connects the third and forth β-strands to their consecutive helices and form a <scene name='69/694225/Small_beta_domain/1'>small β-domain</scene> that consists of a short five-stranded βsheet (β6,β7,β9,β10,β11) that lies on top of the α/β barrel. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> A study of the equilibria between the four subunits shows that each isocitrate lyase monomer has a dynamic comformational change of the active site loop. At any given time, only two of the subunits are in the open conformation. <ref name="gould"> Gould, T.; van de Langemheen, H.; Muñoz-Elías, E.; McKinney, D.; Sacchettini, J.; Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in ''Mycobacterium tuberculosis''. ''Molecular Microbiology''. '''2006'''. ''61(4)'':940-947. doi:10.1111/j.1365-2958.2006.05297.x. </ref> Isocitrate Lyase shows a resemblance to [http://www.rcsb.org/pdb/explore/explore.do?structureId=1S2V phosphoenolpyrvate mutase]. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> | ||
| + | |||
| + | |||
| Line 18: | Line 20: | ||
| - | === | + | |
| + | |||
| + | ===Active Site=== | ||
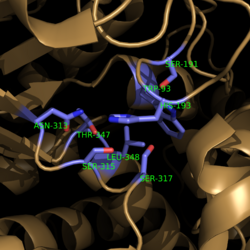
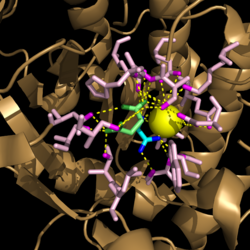
[[Image:Active Site Residues.png|250 px|left|thumb|'''Figure 3. Active Site Residues.''' All eight active site residues necessary for catalysis of isocitrate are shown in slate. However, the protein shown is a C191S mutant of isocitrate lyase.]] [[Image:Active_Site_Hydrogen_Bonding.png|250 px|right|thumb|'''Figure 4. Active site residues hydrogen bound to a cofactor and the products of the catalyzed isocitrate reaction.''' Glyoxylate is shown in blue, succinate is shown in green, and the Mg<sup>2+</sup> cofactor is shown in yellow.]] The active site of isocitrate lyase consists of eight residues: Trp93, Cys191, His193, Ser315, Ser317, Asn313, Thr347, Leu348 (Figure 3). Additionally, there are several other amino acid side chains present that form hydrogen bonding opportunities with isocitrate to catalyze the breakdown reaction to glyoxylate and succinate. Ser91, Gly92, Trp93, and Arg228 form hydrogen bonds with glyoxylate while Asn313, Glu295, Arg228, and Gly192 and Trp93, Thr347, Ser315, Ser317, and His193 form hydrogen bonding opportunities with the two carboxylates within succinate. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> Additionally, a Mg<sup>2+</sup> ion is needed for further electrostatic stabilization of the extreme negative charge on isocitrate. This Mg<sup>2+</sup> hydrogen bonds to the carboxylate in glyoxylate and one of the carboxylates in succinate. | [[Image:Active Site Residues.png|250 px|left|thumb|'''Figure 3. Active Site Residues.''' All eight active site residues necessary for catalysis of isocitrate are shown in slate. However, the protein shown is a C191S mutant of isocitrate lyase.]] [[Image:Active_Site_Hydrogen_Bonding.png|250 px|right|thumb|'''Figure 4. Active site residues hydrogen bound to a cofactor and the products of the catalyzed isocitrate reaction.''' Glyoxylate is shown in blue, succinate is shown in green, and the Mg<sup>2+</sup> cofactor is shown in yellow.]] The active site of isocitrate lyase consists of eight residues: Trp93, Cys191, His193, Ser315, Ser317, Asn313, Thr347, Leu348 (Figure 3). Additionally, there are several other amino acid side chains present that form hydrogen bonding opportunities with isocitrate to catalyze the breakdown reaction to glyoxylate and succinate. Ser91, Gly92, Trp93, and Arg228 form hydrogen bonds with glyoxylate while Asn313, Glu295, Arg228, and Gly192 and Trp93, Thr347, Ser315, Ser317, and His193 form hydrogen bonding opportunities with the two carboxylates within succinate. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> Additionally, a Mg<sup>2+</sup> ion is needed for further electrostatic stabilization of the extreme negative charge on isocitrate. This Mg<sup>2+</sup> hydrogen bonds to the carboxylate in glyoxylate and one of the carboxylates in succinate. | ||
| + | |||
| + | |||
| Line 26: | Line 32: | ||
===Catalytic Loop=== | ===Catalytic Loop=== | ||
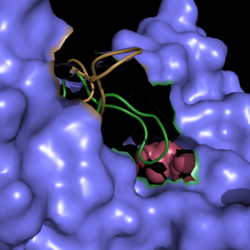
[[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 5. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop unbound is shown in wheat and the active site loop bound is shown in green.]] The catalytic loop of isocitrate lyase consists of residues 185-196. The two most important are Cys191 and His193 as these form a charge relay strong enough to extract a proton from isocitrate. Poor electron density has been observed for residues His193 and Leu194 indicating that this loop is very flexible. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This data backs up the claim that that monomers of the protein are in a structural equilibria between the open and closed forms of the active site. In order for the catalytic loop to shift into the closed position necessary for catalysis, isocitrate must be within the binding pocket. The hydrogen bonding opportunities formed cause a ripple effect that shifts the catalytic loop into a closer position. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This shift also causes the C-terminal domain of the subunit (residues 411-428) to move into the former position of the catalytic loop. The C-terminal domain is stabilized by an electrostatic interaction with Lys189. This combined movement locks the active site residues into a proper orientation for lysis of a C-C bond within isocitrate. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> | [[Image:Active Loop Shift.png|250 px|left|thumb|'''Figure 5. Active Site Loop Shift.''' Binding of the ligand to the enzyme results in a conformational shift that facilitates the breakdown of isocitrate. The active site loop unbound is shown in wheat and the active site loop bound is shown in green.]] The catalytic loop of isocitrate lyase consists of residues 185-196. The two most important are Cys191 and His193 as these form a charge relay strong enough to extract a proton from isocitrate. Poor electron density has been observed for residues His193 and Leu194 indicating that this loop is very flexible. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This data backs up the claim that that monomers of the protein are in a structural equilibria between the open and closed forms of the active site. In order for the catalytic loop to shift into the closed position necessary for catalysis, isocitrate must be within the binding pocket. The hydrogen bonding opportunities formed cause a ripple effect that shifts the catalytic loop into a closer position. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> This shift also causes the C-terminal domain of the subunit (residues 411-428) to move into the former position of the catalytic loop. The C-terminal domain is stabilized by an electrostatic interaction with Lys189. This combined movement locks the active site residues into a proper orientation for lysis of a C-C bond within isocitrate. <ref name="sharma"> Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; ''et. al''; Structure of isocitrate lyase, a persistence factor of ''Mycobacterium tuberculosis''. ''Nat. Struct. Biol.''. '''2000'''. ''7(8)'':663-668. </ref> | ||
| + | |||
| + | |||
| Line 32: | Line 40: | ||
===Regulation=== | ===Regulation=== | ||
Isocitrate lyase competes with [http://en.wikipedia.org/wiki/Isocitrate_dehydrogenase isocitrate dehydrogenase], an enzyme found in the [http://en.wikipedia.org/wiki/Citric_acid_cycle citric cycle], for isocitrate processing. The favoritism of one enzyme over the other is controlled by the phosphorylation of isocitrate dehydrogenase. This enzyme has a much higher affinity for isocitrate as compared to isocitrate lyase. Phosphorylation of isocitrate dehydrogenase inactivates the enzyme and leades to increased isocitrate lyase activity. <ref name="cozzone"> Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. ''Annual Review of Microbiology''. '''1998''', ''52'':127-164. doi: 10.1146/annurev.micro.52.1.127. </ref> | Isocitrate lyase competes with [http://en.wikipedia.org/wiki/Isocitrate_dehydrogenase isocitrate dehydrogenase], an enzyme found in the [http://en.wikipedia.org/wiki/Citric_acid_cycle citric cycle], for isocitrate processing. The favoritism of one enzyme over the other is controlled by the phosphorylation of isocitrate dehydrogenase. This enzyme has a much higher affinity for isocitrate as compared to isocitrate lyase. Phosphorylation of isocitrate dehydrogenase inactivates the enzyme and leades to increased isocitrate lyase activity. <ref name="cozzone"> Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. ''Annual Review of Microbiology''. '''1998''', ''52'':127-164. doi: 10.1146/annurev.micro.52.1.127. </ref> | ||
| + | |||
| + | |||
| Line 41: | Line 51: | ||
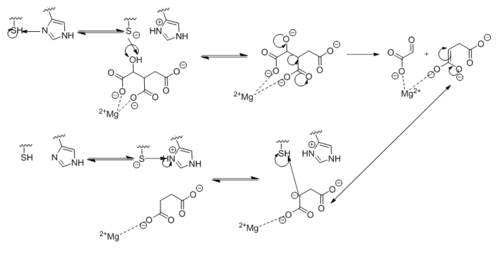
[[Image:Complete_Mechanism.PNG|500 px|left|thumb|'''Figure 7. Observed Mechanism for the Breakdown of Isocitrate by Isocitrate Lyase.''' His193 shifts the pKa of Cys191 and removes its proton. This allows Cys191 to extract a proton from the hydroxyl group of isocitrate. The resulting oxyanion forms a carbonyl and forces the lysis of a C-C bond. Glyoxylate and the enol form of succinate are formed and stabilized with a Mg<sup>2+</sup> ion. The succinate enolate resonates and extracts the proton back from Cys191 to form succinate.]] | [[Image:Complete_Mechanism.PNG|500 px|left|thumb|'''Figure 7. Observed Mechanism for the Breakdown of Isocitrate by Isocitrate Lyase.''' His193 shifts the pKa of Cys191 and removes its proton. This allows Cys191 to extract a proton from the hydroxyl group of isocitrate. The resulting oxyanion forms a carbonyl and forces the lysis of a C-C bond. Glyoxylate and the enol form of succinate are formed and stabilized with a Mg<sup>2+</sup> ion. The succinate enolate resonates and extracts the proton back from Cys191 to form succinate.]] | ||
| + | |||
| + | |||
==Disease Association== | ==Disease Association== | ||
| + | |||
| + | |||
Revision as of 04:40, 10 April 2015
Isocitrate Lyase from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Sharma, V.; Sharma, S.; Hoener zu Bentrup, K.; McKinney, J.; Russell, D.; et. al; Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat. Struct. Biol.. 2000. 7(8):663-668.
- ↑ Gould, T.; van de Langemheen, H.; Muñoz-Elías, E.; McKinney, D.; Sacchettini, J.; Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Molecular Microbiology. 2006. 61(4):940-947. doi:10.1111/j.1365-2958.2006.05297.x.
- ↑ Cozzone, A.; Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annual Review of Microbiology. 1998, 52:127-164. doi: 10.1146/annurev.micro.52.1.127.
- ↑ Srivastava, V.; Janin, A.; Srivastava, B.; Srivastava, R.; Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. ScienceDirect. 2007. doi:10.1016/j.tube.2007.10.002.
- ↑ Muñoz-Elías, E.; McKinney, J.; M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005. 11(6):638-644. doi:10.1038/nm1252.
- ↑ Dunn, M.; Ramírez-Trujillo, J.; Hernández-Lucas, I.; Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009. 155:3166-3175. doi:10.1099/mic.0.030858-0.