Sandbox Reserved 1061
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Background == | == Background == | ||
| - | ''Mycobacterium tuberculosis'' [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Wikipedia]resides in the lungs of a host and upon becoming active, results in symptoms such as chest pains, weakness, and intense coughing. Left untreated and unmanaged, TB can lead to death (1.5 million in 2013). The disease has a high co-morbidity with HIV/AIDS due to its immunocompromising tendencies. Tuberculosis is one of the most heavily studied diseases today. With over 9 million infections worldwide per year, the necessity for antimicrobial agents to combat emerging multi-drug resistant strands is imperative. | + | ''Mycobacterium tuberculosis'' [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Wikipedia]resides in the lungs of a host and upon becoming active, results in symptoms such as chest pains, weakness, and intense coughing. Left untreated and unmanaged, TB can lead to death (1.5 million in 2013).<ref>"Tuberculosis." Media Centre. World Health Organization, Web. 16 Mar. 2015. Media Centre. <http://www.who.int/mediacentre/factsheets/fs104/en/>.</ref>The disease has a high co-morbidity with HIV/AIDS due to its immunocompromising tendencies. Tuberculosis is one of the most heavily studied diseases today. With over 9 million infections worldwide per year, the necessity for antimicrobial agents to combat emerging multi-drug resistant strands is imperative. |
| + | |||
== Structure == | == Structure == | ||
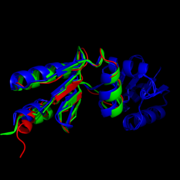
| - | The structure of ''M. tuberculosis'' as determined by x-ray crystallography has 79 residues in a single polypeptide chain. <scene name='69/694228/Nrdh_structure/1'>NrdH Chrystal Structure</scene>. The active site (shown in green) is dominated by a disulphide bond between Cys-11 and Cys-14, which serves as the site of reduction by Theirodoxin reductase. | + | The structure of ''M. tuberculosis'' as determined by x-ray crystallography has 79 residues in a single polypeptide chain. <scene name='69/694228/Nrdh_structure/1'>NrdH Chrystal Structure</scene>. The active site (shown in green) is dominated by a disulphide bond between Cys-11 and Cys-14, which serves as the site of reduction by Theirodoxin reductase. <ref>Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.</ref> |
| - | Many theirodoxin-like proteins have a similar active site region, denoted as the theirodoxin fold, which occurs directly before the disulfide bond. The residues in this region, denoted by letters CVQC, are the most highly conserved of all areas of the protein across multiple species. Exactly how this structure relates to function is somewhat debated. A Threonine-7 reside directly across the theirodoxin fold from the disulphide bond has been suggested to adopt two different conformations which differentially affect the redox abilities of the Protein. In the <scene name='69/694228/Nrdh_ligand_binding_site/8'>"A" Conformation</scene>, the alcohol of the threonine side chain points towards the disulfide bond, engaging an ionic interaction between the two that prevents the Therodoxin Reductase from binding. Alternatively, in the <scene name='69/694228/Nrdh_ligand_binding_site/12'>"B" Conformation</scene>, the alcohol points in the opposite direction, allowing sufficient space for the ligand to bind and reduction to occur. | + | Many theirodoxin-like proteins have a similar active site region, denoted as the theirodoxin fold, which occurs directly before the disulfide bond. The residues in this region, denoted by letters CVQC, are the most highly conserved of all areas of the protein across multiple species. Exactly how this structure relates to function is somewhat debated. A Threonine-7 reside directly across the theirodoxin fold from the disulphide bond has been suggested to adopt two different conformations which differentially affect the redox abilities of the Protein. In the <scene name='69/694228/Nrdh_ligand_binding_site/8'>"A" Conformation</scene>, the alcohol of the threonine side chain points towards the disulfide bond, engaging an ionic interaction between the two that prevents the Therodoxin Reductase from binding. Alternatively, in the <scene name='69/694228/Nrdh_ligand_binding_site/12'>"B" Conformation</scene>, the alcohol points in the opposite direction, allowing sufficient space for the ligand to bind and reduction to occur.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.</ref> |
| - | The active site of the protein is stabilized through a network of hydrogen bonds involving the two highly conserved residues, CVQC and WSGFRP. The crystal structure shows that interactions with one water molecule is necessary for the proper coordination between the conserved motifs to occur. These hydrogen bonds orient the important residues in the most optimal position to promote oxidation and reduction. | + | The active site of the protein is stabilized through a network of hydrogen bonds involving the two highly conserved residues, CVQC and WSGFRP. The crystal structure shows that interactions with one water molecule is necessary for the proper coordination between the conserved motifs to occur. These hydrogen bonds orient the important residues in the most optimal position to promote oxidation and reduction.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.</ref> |
[[Image:Weblogocvqc.png|thumb|center|upright=2.5|Weblogo diagram showing highly conserved CVQC region of NrdH.]] | [[Image:Weblogocvqc.png|thumb|center|upright=2.5|Weblogo diagram showing highly conserved CVQC region of NrdH.]] | ||
| Line 36: | Line 37: | ||
Like most NrdHs, MtNrdH is similar in sequence to glutaredoxins, but structurally similar to thioredoxins. MtNrdH also accepts electrons from thiordoxin reductase, a characteristic of thiordoxins, but not glutaredoxins.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4057.</ref> [[Image:Image-Super imposed molecules.png|thumb|left|Structural comparison of NrdHs with "thioredoxin folds": ''E. Coli'' NrdH (green), ''C. ammoniagenes'' NrdH (blue), ''M. tuberculosis'' NrdH (red)]]<ref>DOI 10.1002/ijch.201300024</ref> <ref>PMID:21638687</ref> | Like most NrdHs, MtNrdH is similar in sequence to glutaredoxins, but structurally similar to thioredoxins. MtNrdH also accepts electrons from thiordoxin reductase, a characteristic of thiordoxins, but not glutaredoxins.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4057.</ref> [[Image:Image-Super imposed molecules.png|thumb|left|Structural comparison of NrdHs with "thioredoxin folds": ''E. Coli'' NrdH (green), ''C. ammoniagenes'' NrdH (blue), ''M. tuberculosis'' NrdH (red)]]<ref>DOI 10.1002/ijch.201300024</ref> <ref>PMID:21638687</ref> | ||
| - | Similar structures of NrdH have been isolated in other primitive species including ''E. coli'', ''S. pyogenes'', ''S. typhimurium'', ''D. deserti'', ''S. flexneri 2457T'', and ''S. dysenteriae''. In higher order multi-cellular organisms, however the NrdH protein is replaced by more complex glutaredoxins or theirodoxins. This observation leads some to speculate that NrdH is one of the very first ancestors in the ribonucleotide reduction pathway. If this is true, NrdH can be seen as a critical protein that allowed for the development of DNA-based life since deoxyribonucleotides could not have existed without the ribonucleotide reduction pathway. A better understanding of the evolutionary timeline of NrdH and similar proteins could shed greater light onto the RNA Wold Hypothesis, specifically describing the time frame of emergence of DNA based life. | + | Similar structures of NrdH have been isolated in other primitive species including ''E. coli'', ''S. pyogenes'', ''S. typhimurium'', ''D. deserti'', ''S. flexneri 2457T'', and ''S. dysenteriae''. In higher order multi-cellular organisms, however the NrdH protein is replaced by more complex glutaredoxins or theirodoxins. This observation leads some to speculate that NrdH is one of the very first ancestors in the ribonucleotide reduction pathway. <ref>Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.</ref> If this is true, NrdH can be seen as a critical protein that allowed for the development of DNA-based life since deoxyribonucleotides could not have existed without the ribonucleotide reduction pathway. A better understanding of the evolutionary timeline of NrdH and similar proteins could shed greater light onto the RNA Wold Hypothesis, specifically describing the time frame of emergence of DNA based life. |
Revision as of 17:25, 10 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Structure of Mycobacterium Tuberculosis NrdH
| |||||||||||
References
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ "Tuberculosis." Media Centre. World Health Organization, Web. 16 Mar. 2015. Media Centre. <http://www.who.int/mediacentre/factsheets/fs104/en/>.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4060.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4057.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4056.
- ↑ Nelson, David L., and Michael M. Cox. Lehninger Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. 888-889.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4056.
- ↑ Makhlynets, O., Boal, A. K., Rhodes, D. V., Kitten, T., Rosenzweig, A. C., & Stubbe, J. (2014). Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS. The Journal of Biological Chemistry, 289(9), 6259–6272. doi:10.1074/jbc.M113.533554.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4057.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644