We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1061
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
== Function == | == Function == | ||
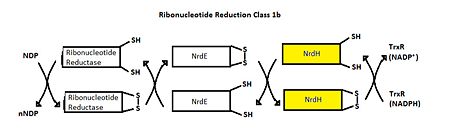
| - | MtNrdH has been identified as an electron carrier protein in ribonuleotide reduction. Ribonucleotide reduction uses an enzyme called ribonucleotide reductase (RNR) to make deoxyribonucleotides, which act as precursors to DNA synthesis. Three classes of RNRs have been identified; each class differs in cofactor requirement, structure, and oxygen dependence, but the general catalytic mechanism is conserved in all three classes.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4056.</ref> Mycobacterium tuberculosis uses class I ribonucleotide reductase. | + | MtNrdH has been identified as an electron carrier protein in ribonuleotide reduction. Ribonucleotide reduction uses an enzyme called [http://www.proteopedia.org/wiki/index.php/Ribonucleotide_reductase ribonucleotide reductase (RNR)] to make deoxyribonucleotides, which act as precursors to DNA synthesis. Three classes of RNRs have been identified; each class differs in cofactor requirement, structure, and oxygen dependence, but the general catalytic mechanism is conserved in all three classes.<ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4056.</ref> Mycobacterium tuberculosis uses class I ribonucleotide reductase. |
Class I RNR is further subdivided into class Ia and Ib. Both Ia and Ib reduce ribonucleotide 5’ diphosphate to deoxyribonucleotide 5’ diphosphate (NDP to dNDP). After ribonucleotide reductase performs the first round of reduction, RNR must be reduced again to reset the cycle. In class Ib, RNR is reduced by either glutadoxin or thiordoxin, which are first reduced by glutadoxin reductase and thiordoxin reductase, respectively.<ref>Nelson, David L., and Michael M. Cox. Lehninger Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. 888-889.</ref> In class Ib, RNR is reduced by NrdE, which is first reduced by NrdH. An important distinction between Ia and Ib is that Ia is present in eukaryotes, eubacteria, bacteriophages, and virus, but Ib is only present in eubacteria. <ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4056.</ref> | Class I RNR is further subdivided into class Ia and Ib. Both Ia and Ib reduce ribonucleotide 5’ diphosphate to deoxyribonucleotide 5’ diphosphate (NDP to dNDP). After ribonucleotide reductase performs the first round of reduction, RNR must be reduced again to reset the cycle. In class Ib, RNR is reduced by either glutadoxin or thiordoxin, which are first reduced by glutadoxin reductase and thiordoxin reductase, respectively.<ref>Nelson, David L., and Michael M. Cox. Lehninger Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. 888-889.</ref> In class Ib, RNR is reduced by NrdE, which is first reduced by NrdH. An important distinction between Ia and Ib is that Ia is present in eukaryotes, eubacteria, bacteriophages, and virus, but Ib is only present in eubacteria. <ref>Swastik, Phulera and Mande, Shekhar C. (2013) 4056.</ref> | ||
Revision as of 19:03, 10 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Structure of Mycobacterium Tuberculosis NrdH
| |||||||||||
References
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ "Tuberculosis." Media Centre. World Health Organization, Web. 16 Mar. 2015. Media Centre. <http://www.who.int/mediacentre/factsheets/fs104/en/>.
- ↑ "Tuberculosis." Media Centre. World Health Organization, Web. 16 Mar. 2015. Media Centre. <http://www.who.int/mediacentre/factsheets/fs104/en/>.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4060.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4057.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4056.
- ↑ Nelson, David L., and Michael M. Cox. Lehninger Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. 888-889.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4056.
- ↑ Makhlynets, O., Boal, A. K., Rhodes, D. V., Kitten, T., Rosenzweig, A. C., & Stubbe, J. (2014). Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS. The Journal of Biological Chemistry, 289(9), 6259–6272. doi:10.1074/jbc.M113.533554.
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) 4057.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644