Sandbox Reserved 1070
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
This domain of MgtC, in contrast, is highly variable in comparison to several orthologues, as presented by Yang et al. However, through a sequence alignment of five known functional MgtC orthologues from pathogens that survive inside macrophages (''M. tuberculosis, B. melitensis, B. cenocepacia, Y. pestis,'' and ''S. Typhimurium''), seven strictly conserved residues were found to be scattered along the whole sequence of the relatively hydrophilic and soluble C-terminal domain. | This domain of MgtC, in contrast, is highly variable in comparison to several orthologues, as presented by Yang et al. However, through a sequence alignment of five known functional MgtC orthologues from pathogens that survive inside macrophages (''M. tuberculosis, B. melitensis, B. cenocepacia, Y. pestis,'' and ''S. Typhimurium''), seven strictly conserved residues were found to be scattered along the whole sequence of the relatively hydrophilic and soluble C-terminal domain. | ||
| - | A large hydrophobic core has conserved residues <scene name='69/698113/Conserved_core_residues/ | + | A large hydrophobic core has conserved residues <scene name='69/698113/Conserved_core_residues/3'>Cysteine-155, Arginine-164, Glutamine-160, and Alanine-195</scene>. |
| + | |||
| + | [[Image:Core_Web_Logo.PNG |600 × 155 px|thumb|left|Four strictly conserved residues of five known functional MgtC orthologs of the soluble C-terminal domain. | ||
| + | The figure was prepared using the WebLogo. (http://weblogo.berkeley.edu/)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
The opposite side of the protein has a small cluster of conserved residues <scene name='69/698113/Conserved_surface_residues/1'>Tyrosine-149, Glutamine-208, and Tryptophan-225</scene>. | The opposite side of the protein has a small cluster of conserved residues <scene name='69/698113/Conserved_surface_residues/1'>Tyrosine-149, Glutamine-208, and Tryptophan-225</scene>. | ||
| - | [[Image: | + | [[Image:Surface_Web_Logo.PNG |419 × 150 px|thumb|left|Four strictly conserved residues of five known functional MgtC orthologs of the soluble C-terminal domain.]] |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
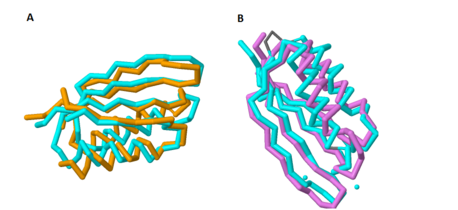
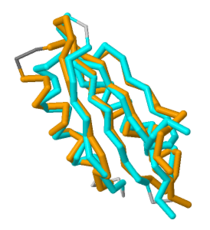
Additionally, there is a crystal structure available for this domain. When comparing the crystal structure of the C-terminal domain to other protein structures, there are striking similarities between this domain and a class of proteins known as ACT domains. | Additionally, there is a crystal structure available for this domain. When comparing the crystal structure of the C-terminal domain to other protein structures, there are striking similarities between this domain and a class of proteins known as ACT domains. | ||
| Line 53: | Line 103: | ||
==Future Work== | ==Future Work== | ||
[[Image:Aligned 3.png|210 px|thumb|right|'''Figure 2. Overlap of MgtC C-terminal domain with the ACT domain of a GTP pyrophosphokinase.''' This figure demonstrates the significant overlap between the C-terminal domain of MgtC and the ACT domain of a GTP pyrophosphokinase.]] | [[Image:Aligned 3.png|210 px|thumb|right|'''Figure 2. Overlap of MgtC C-terminal domain with the ACT domain of a GTP pyrophosphokinase.''' This figure demonstrates the significant overlap between the C-terminal domain of MgtC and the ACT domain of a GTP pyrophosphokinase.]] | ||
| - | Since so little is known about MgtC, future work should involve both crystallizing the entire MgtC protein and characterizing its biochemical function. Because the sequence of amino acids in a protein dictates structure, and structure typically determines the protein's function, further sequencing and structural analysis should be performed with MgtC to discern its function. | + | Since so little is known about MgtC, future work should involve both crystallizing the entire MgtC protein and characterizing its biochemical function. Because the sequence of amino acids in a protein dictates structure, and structure typically determines the protein's function, further sequencing and structural analysis should be performed with MgtC to discern its function. Shown in '''Figure 2''' is an overlap of MgtC with the ACT domain of a GTP pyrophosphokinase. This overlap shows even more extensive similarity than the aforementioned SerA and NikR ACT domains. Structural similarity analysis could aid in resolving the biochemical function of MgtC. |
Revision as of 22:14, 10 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
MgtC: A Virulence Factor From Mycobacterium tuberculosis
| |||||||||||