Sandbox Reserved 1068
From Proteopedia
| Line 45: | Line 45: | ||
'''Isochorismate pyruvae lyase (IPL)''' | '''Isochorismate pyruvae lyase (IPL)''' | ||
| + | |||
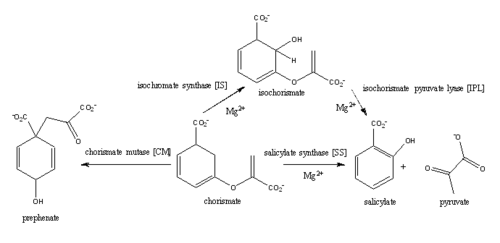
For IPL activity, a proposed histidine residue (his334) acts as a base, abstracting the C2 proton of isochorismate through a second order elimination mechanism. The C9 atom of isochorismate is then protonated, leading to the elimination of pyruvate and formation of salicylate. At pH values below 7.5 the histidine residue is protonated and therefore not able to act as a base in the mechanism, causing an accumulation of isochorismate. | For IPL activity, a proposed histidine residue (his334) acts as a base, abstracting the C2 proton of isochorismate through a second order elimination mechanism. The C9 atom of isochorismate is then protonated, leading to the elimination of pyruvate and formation of salicylate. At pH values below 7.5 the histidine residue is protonated and therefore not able to act as a base in the mechanism, causing an accumulation of isochorismate. | ||
'''Isochorismate synthast (IS)''' | '''Isochorismate synthast (IS)''' | ||
| + | |||
The C1 carboxylate group of chorismate binds to the magnesium cation within the active site. | The C1 carboxylate group of chorismate binds to the magnesium cation within the active site. | ||
| - | Currently, isochorismate is believed to be formed from chorismate through a proposed Sn2 mechanism involving nucleophilic attack of an activated water molecule to the C2 center followed by either a concerted or stepwise elimination of the C4 hydroxyl group'''(He et al.)'''. Lys205 has been proposed to act as the catalytic base, activating a water molecule in the active site by abstracting one of its protons. However, mutational analysis of Lys205 suggested that the lysine reside is not the sole determinant in the activation of a water molecule for nucleophilic attack of the C2 center. Studies have shown that Lys205 is protonated at neutral pH and therefore can't act as a base to activate the water molecule, agreeing with the mutational analysis data. Instead of Lys205, Glu297 residue has been proposed to act as a base in the activation of the water molecule. | + | Currently, isochorismate is believed to be formed from chorismate through a proposed Sn2 mechanism involving nucleophilic attack of an activated water molecule to the C2 center followed by either a concerted or stepwise elimination of the C4 hydroxyl group'''(He et al.)'''. Lys205 has been proposed to act as the catalytic base, activating a water molecule in the active site by abstracting one of its protons. However, mutational analysis of Lys205 suggested that the lysine reside is not the sole determinant in the activation of a water molecule for nucleophilic attack of the C2 center. Studies have shown that Lys205 is protonated at neutral pH and therefore can't act as a base to activate the water molecule, agreeing with the mutational analysis data. The magnesium ion also causes the Lys205 residue is also displaced away from the active site. Instead of Lys205, Glu297 residue has been proposed to act as a base in the activation of the water molecule. The magnesium ion forces the negatively charged Glu297 residue to face toward the active site and the pKa of Glu297 (3.9) suggest an unprotonated state. Furthermore, Glu297 forms a hydrogen bond with a water molecule within the active site as well as with Lys205, which is in turn hydrogen bonded to C1 carboxylate group of chorismate and the oxygen of the nucleophilic water molecule. The glutamic residue (Gly252) could protonate the C4 leaving hydroxyl group. The pKa of Gly252 (7.7) suggest that is it is the only protonated glutamate residue in the active site at pH 7 and thus able to protonate the C4 leaving group. The pKa of Gly252 also accounts for the accumulation of isochorismate at pH values below 7.5. |
'''isochorismate pyruvate lyase (IPL)''' | '''isochorismate pyruvate lyase (IPL)''' | ||
Revision as of 00:59, 11 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Mycobacterium tuberculosis salicylate synthase (Mbt1)
| |||||||||||
References
1. Chi G1, Manos-Turvey A, O'Connor PD, Johnston JM, Evans GL, Baker EN, Payne RJ, Lott JS, Bulloch EM. 2012. Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis. Biochemistry 51(24):4868-79. doi: 10.1021/bi3002067
2. Ferrer S1, Martí S, Moliner V, Tuñón I, Bertrán J. 2012 Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 14(10):3482-9. doi: 10.1039/c2cp23149b.
3. Harrison AJ1, Yu M, Gårdenborg T, Middleditch M, Ramsay RJ, Baker EN, Lott JS. 2006. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J Bacteriol. 188(17):6081-91.
4. Manos-Turvey A1, Cergol KM, Salam NK, Bulloch EM, Chi G, Pang A, Britton WJ, West NP, Baker EN, Lott JS, Payne RJ. 2012. Synthesis and evaluation of M. tuberculosis salicylate synthase (MbtI) inhibitors designed to probe plasticity in the active site. Org Biomol Chem 10(46):9223-36. doi: 10.1039/c2ob26736e.
5. Zwahlen J1, Kolappan S, Zhou R, Kisker C, Tonge PJ. 2007. Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 46(4):954-64.