Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
== Structural Highlights == | == Structural Highlights == | ||
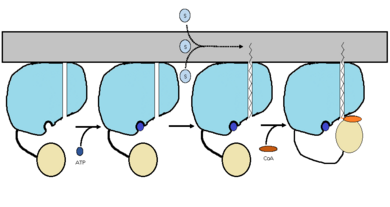
FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a peripheral-membrane proteins. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. There are two domains of this protein, a larger N-terminal domain (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and a smaller C-terminal domain (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>). These domains are held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>). Inside the larger N-terminal domain is a hydrophobic tunnel, which allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine-rich lid loop that is involved in the peripheral binding of the enzyme to the membrane<ref> [http://www.proteopedia.org/wiki/index.php/3r44/ 3R44] </ref>. Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell wall. | FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a peripheral-membrane proteins. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. There are two domains of this protein, a larger N-terminal domain (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and a smaller C-terminal domain (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>). These domains are held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>). Inside the larger N-terminal domain is a hydrophobic tunnel, which allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine-rich lid loop that is involved in the peripheral binding of the enzyme to the membrane<ref> [http://www.proteopedia.org/wiki/index.php/3r44/ 3R44] </ref>. Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell wall. | ||
| - | |||
| - | The first important structural point of note is that there are two subunits, the larger N-terminal subunit (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and the smaller C-terminal subunit (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>) held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>).The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. The next major region of note is the hydrophobic tunnel which allows for lipid binding up to 26 carbons in length and extends from the ATP and AMP binding region. The hydrophobic tunnel is capped by an arginine and aromatic-rich surface patch which is involved in membrane binding of the protein<ref> [http://www.proteopedia.org/wiki/index.php/3r44/ 3R44] </ref>.Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell wall. | ||
== Function == | == Function == | ||
Revision as of 12:54, 14 April 2015
FadD13
| |||||||||||