We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
== Structural Highlights == | == Structural Highlights == | ||
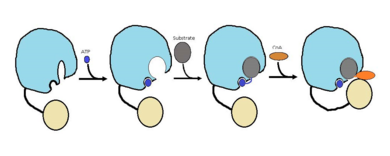
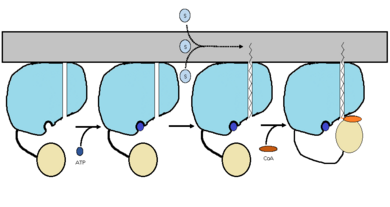
| - | FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a [http://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral-membrane protein]. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. There are two domains of this protein, a larger N-terminal domain (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and a smaller C-terminal domain (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>). These domains are held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>). Inside the larger N-terminal domain is a hydrophobic tunnel, which allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine-rich lid loop that is involved in the peripheral binding of the enzyme to the membrane. Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell | + | FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a [http://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral-membrane protein]. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The <scene name='69/694230/Atp_and_amp_binding_region/4'>ATP and AMP binding region</scene> allows for either ATP or AMP to bind and activate FadD13. There are two domains of this protein, a larger N-terminal domain (<scene name='69/694230/N-terminal_domain/5'>residues 1-395</scene>) and a smaller C-terminal domain (<scene name='69/694230/C-terminal_domain/1'>residues 402-503</scene>). These domains are held together by a six amino acid linker (<scene name='69/694230/Residues_396-401/1'>residues 396-401</scene>). Inside the larger N-terminal domain is a hydrophobic tunnel, which allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine-rich lid loop that is involved in the peripheral binding of the enzyme to the membrane. Three key arginine residues, <scene name='69/694230/Arginine_surface_patch/1'>Arg 195, 197, and 199</scene> all play an important role for the enzyme to be able to bind to the cell membrane. |
== Function == | == Function == | ||
Revision as of 13:34, 14 April 2015
FadD13
| |||||||||||
References
- ↑ Watkins, P.A., Maiguel, D., Jia, Z., and Pevsner, J. (2007). Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48, 2736-2750.
- ↑ Kochan, G., Pilka, E.S., von Delft, F., Oppermann, U., and Yue,W.W. (2009). Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl0coenzyme A synthetase ACSM2A. J. Mol. Biol. 388, 997-1008.
- ↑ Andersson, C.S., Lundgren, C.A.K., Magnusdottir, A., Ge, C., Weislander, A., Molina, D., Hogbom, M. (2012)The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: structural Basis for Housing lipid Substrates longer than the Enzyme. Cell Press,1062-1070