This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1074
From Proteopedia
| Line 51: | Line 51: | ||
=== Isoniazid === | === Isoniazid === | ||
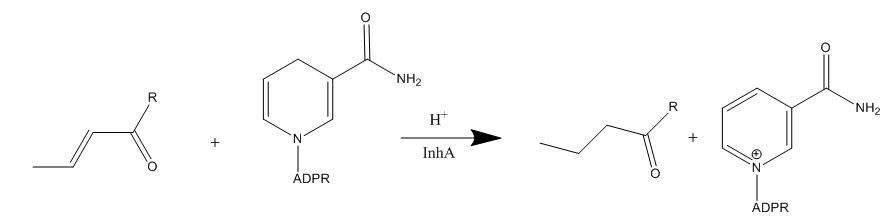
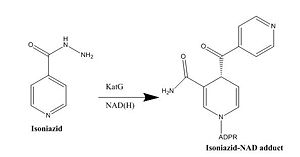
| - | Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser-94 to Ala of InhA was sufficient enough to have isoniazid resistance <ref name="InhA" />. The mutation of Ser-94 to alanine is found around the cofactor binding site, resulting in a 20 to 160-fold reduction in affinity of NADH for the enzyme <ref name=”Ser94”> Kruh, N. “et al.” (2007). Probing mechanisms of resistance to the tuberculosis drug isoniazid: Conformational changes caused by inhibition of InhA, the enoyl reductase from | + | Isoniazid is a first-line antibiotic that has been used to treat tuberculosis infections for over 50 years. Isoniazid is known to inhibit mycolic acid biosnthesis, which is the function of InhA. The activated form of isoniazid is covalently attached to the [http://en.wikipedia.org/wiki/Nicotinamide nicotinamide] ring of NADH. However, Isoniazid is still not an ideal antibiotic because many drug-resistant strains of tuberculosis have shown resistance to this inhibitor. Specifically, the mutation Ser-94 to Ala of InhA was sufficient enough to have isoniazid resistance <ref name="InhA" />. The mutation of Ser-94 to alanine is found around the cofactor binding site, resulting in a 20 to 160-fold reduction in affinity of NADH for the enzyme <ref name=”Ser94”> Kruh, N. “et al.” (2007). Probing mechanisms of resistance to the tuberculosis drug isoniazid: Conformational changes caused by inhibition of InhA, the enoyl reductase from ''Mycobacterium tuberculosis''. ''Protein Sci, 16(8),'' 1617-1627. PMID: [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2203378/ 17600151] </ref>. |
=== Other Inhibitors === | === Other Inhibitors === | ||
Revision as of 19:57, 15 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Contents |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Bell, A.F. et al.(2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. Journal of the American Chemical Society, 129, 6425-6431. DOI: 10.1021/ja068219m
- ↑ 2.0 2.1 Bhatt, A. et al. (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Journal of Molecular Microbiology, 64(6), 1442-1454. PMID: 17555433 DOI: 10.1111/j.1365-2958.2007.05761.x
- ↑ Marrakchi, Hedia, et al. (2000). InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Journal of Microbiology, 146, 289-296. PMID: 10708367
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Rozwarski, D.A. et al. (1999). Crystal Structure of the Mycobacterium tuberculosis Enoyl-ACP Reductase, InhA, in Complex with NAD+ and a C16 Fatty Acyl Substrate. Journal of Biological Chemistry, 274(22), 15582-15589. PMID: 10336454 DOI: 10.1074/jbc.274.22.15582
- ↑ Kruh, N. “et al.” (2007). Probing mechanisms of resistance to the tuberculosis drug isoniazid: Conformational changes caused by inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis. Protein Sci, 16(8), 1617-1627. PMID: 17600151
- ↑ 6.0 6.1 6.2 Vilchèze, C. et al. (2011). Novel Inhibitors of InhA Efficiently Kill Mycobacterium tuberculosis under Aerobic and Anaerobic Conditions. Antimicrobial Agents and Chemotherapy, 55(8), 3889-3898. DOI: 10.1128/AAC.00266-11
Student Contributors
- Arielle Russell
- Mackenzie A. Smith
Similar Proteopedia Pages
Enoyl-Acyl-Carrier Protein Reductase
Additional 3D Structures of Enoyl-ACP Reductase InhA
3oew, 2x22, 2x23, 1eny, 1enz, 4dqu, 4dre - MtENR+NAD; 3of2, 4dti - MtENR(mutant)+NAD; 2pr2, 2idz, 2h9i - MtENR+INH-NAPD; 2aq8 - MtENR+NADH; 2aqh, 2aqi, 2aqk, 3oey - MtENR(mutant)+NADH; 2ntj - MtENR+PTH-NAD; 2ie0, 2ieb, 2nv6, 1zid - MtENR(mutant)+INH-NAPD; 3fne, 3fnf, 3fng, 3fnh, 2b35, 1p45 - MtENR+NAD+TCI; 2b36, 2b37, 4ohu, 4oim, 4oyr - MtENR+NAD+phenoxyphenol derivative; 2nsd - MtENR+NAD+piperidine derivative; 2h7l, 2h7m, 2h7n, 2h7p, 4u0j, 4tzt, 4tzk, 4trj, 4u0k - MtENR+NAD+pyrrolidine derivative; 4cod, 4bqp, 4bqr, 4bge, 4bii, 4oxk, 4oxn, 4oxy, 4r9r, 4r9s - MtENR+NAD + inhibitor; 4bgi - MtENR (mutant)+NAD+inhibitor; 1p44 - MtENR+NAD+indole derivative; 1bvr - MtENR+NAD+fatty-acyl substrate