Sandbox Reserved 1074
From Proteopedia
| Line 13: | Line 13: | ||
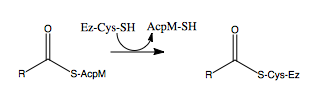
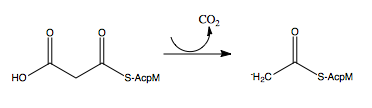
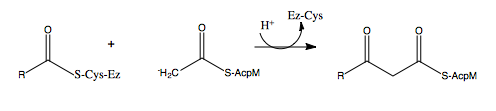
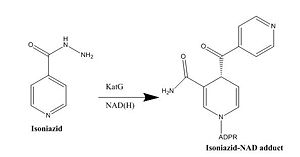
[http://en.wikipedia.org/wiki/Mycolic_acid Mycolic acids] are very long-chain fatty acids (C<sub>60</sub> -C<sub>90</sub>) that are essential components of the mycobacterial cell wall. Mycolic acids are synthesized by at least two known elongation systems, type I and type II [http://en.wikipedia.org/wiki/Fatty_acid_synthase fatty acid synthases] (FAS-I and FAS-II) <ref name="FAS-II"> Bhatt, A. ''et al.'' (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. ''Journal of Molecular Microbiology, 64(6),'' 1442-1454. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/17555433 17555433] DOI: [http://www.ncbi.nlm.nih.gov/pubmed/17555433 10.1111/j.1365-2958.2007.05761.x]</ref>. The FAS-II system prefers C16 as a starting substrate and can extend up to C56 <ref name="FAS-II system"> Marrakchi, Hedia, ''et al.'' (2000). InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. ''Journal of Microbiology, 146,'' 289-296. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/10708367 10708367] </ref>. The FAS-II system utilizes the products from the FAS-I system as primers to extend the chain lengths further. The products of the FAS-II system are the precursors of mycolic acids. Elongation by the FAS-II system occurs by a [http://en.wikipedia.org/wiki/Condensation_reaction condensation reaction]of [http://en.wikipedia.org/wiki/Acetyl acetyl] and [http://en.wikipedia.org/wiki/Malonyl-CoA malonyl] substrates, which is achieved in three steps. Step 1 involves transfer of the acyl primer, step 2 involves [http://en.wikipedia.org/wiki/Decarboxylation decarboxylation] of the substrate to yield a [http://en.wikipedia.org/wiki/Carbanion carbanion], and step 3 involves nucleophilic attack of the carbanion to yield the elongated product <ref name="FAS-II" />. | [http://en.wikipedia.org/wiki/Mycolic_acid Mycolic acids] are very long-chain fatty acids (C<sub>60</sub> -C<sub>90</sub>) that are essential components of the mycobacterial cell wall. Mycolic acids are synthesized by at least two known elongation systems, type I and type II [http://en.wikipedia.org/wiki/Fatty_acid_synthase fatty acid synthases] (FAS-I and FAS-II) <ref name="FAS-II"> Bhatt, A. ''et al.'' (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. ''Journal of Molecular Microbiology, 64(6),'' 1442-1454. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/17555433 17555433] DOI: [http://www.ncbi.nlm.nih.gov/pubmed/17555433 10.1111/j.1365-2958.2007.05761.x]</ref>. The FAS-II system prefers C16 as a starting substrate and can extend up to C56 <ref name="FAS-II system"> Marrakchi, Hedia, ''et al.'' (2000). InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. ''Journal of Microbiology, 146,'' 289-296. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/10708367 10708367] </ref>. The FAS-II system utilizes the products from the FAS-I system as primers to extend the chain lengths further. The products of the FAS-II system are the precursors of mycolic acids. Elongation by the FAS-II system occurs by a [http://en.wikipedia.org/wiki/Condensation_reaction condensation reaction]of [http://en.wikipedia.org/wiki/Acetyl acetyl] and [http://en.wikipedia.org/wiki/Malonyl-CoA malonyl] substrates, which is achieved in three steps. Step 1 involves transfer of the acyl primer, step 2 involves [http://en.wikipedia.org/wiki/Decarboxylation decarboxylation] of the substrate to yield a [http://en.wikipedia.org/wiki/Carbanion carbanion], and step 3 involves nucleophilic attack of the carbanion to yield the elongated product <ref name="FAS-II" />. | ||
| - | [[Image:Screen_Shot_2015-04-15_at_4.07.06_PM.png|thumb| | + | [[Image:Screen_Shot_2015-04-15_at_4.07.06_PM.png|thumb|2000 px|center|Figure 1. Step 1 - Acyl Transfer <ref name="FAS-II"/>]] [[Image:Step.png|thumb|2000 px|center|Figure 2. Step 2 - Decarboxylation <ref name="FAS-II"/>]] |
| - | [[Image:Step_3.png|thumb| | + | [[Image:Step_3.png|thumb|3000 px|center|Figure 3. Step 3 - Condensation <ref name="FAS-II"/>]] |
| Line 46: | Line 46: | ||
==='''Importance of Tyr-158'''=== | ==='''Importance of Tyr-158'''=== | ||
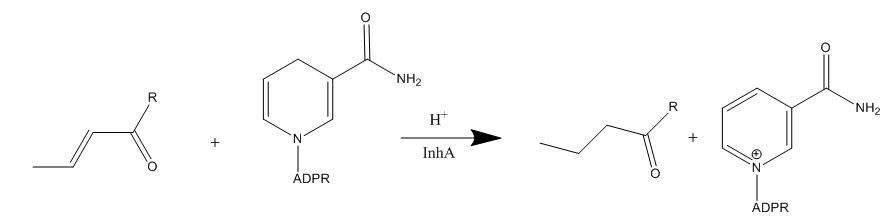
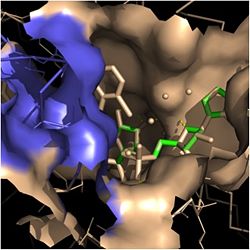
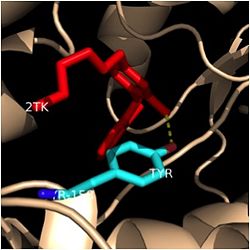
| - | [[Image:Tyr-158.jpg|thumb| | + | [[Image:Tyr-158.jpg|thumb|250px|left|Figure 4. Tyr-158 (light blue) hydrogen bonding to the fatty acyl substrate, 2TK (red)]] One example of an amino acid that is not a part of the substrate binding loop yet interacts with the fatty acyl substrate is Tyr-158. This amino acid is conserved in other enoyl-ACP reductases in both bacteria and plants, so it likely plays an essential role in the function of these specific enzymes. Studies have shown that <scene name='69/694241/Tyr158_2tk/1'>Tyr-158</scene> forms the only direct hydrogen bond that exists between the InhA protein and the fatty acyl substrate. This hydrogen bond occurs between the hydroxyl oxygen on the side chain of Tyr-158 and the thioester carbonyl oxygen of the fatty acyl substrate. Consequently, the Tyr-158 residue acts to stabilize the enolate intermediate that forms during the hydride transfer reaction <ref name="InhA" />. |
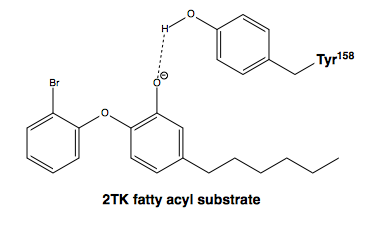
| - | [[Image:Tyr-158_and_2TK.png|thumb|600 px|center|Figure #. The side chain hydroxyl of Tyr-158 stabilizes the enolate intermediate of the fatty acyl substrate (2TK) <ref name=”Tyr-158”> Li, H.J. “et al.” (2014). A Structural and Energetic Model for the Slow-Onset Inhibition of the “Mycobacterium tuberculosis“ Enoyl-ACP Reductase InhA. “ACS Chemical Biology 9,” 986-993. PMID: [http://pubs.acs.org/doi/pdf/10.1021/cb400896g 24527857] </ref>.]] | ||
| + | [[Image:Tyr-158_and_2TK.png|thumb|600 px|center|Figure #. The side chain hydroxyl of Tyr-158 stabilizes the enolate intermediate of the fatty acyl substrate (2TK) <ref name=”Tyr-158”> Li, H.J. “et al.” (2014). A Structural and Energetic Model for the Slow-Onset Inhibition of the “Mycobacterium tuberculosis“ Enoyl-ACP Reductase InhA. “ACS Chemical Biology 9,” 986-993. PMID: [http://pubs.acs.org/doi/pdf/10.1021/cb400896g 24527857] </ref>.]] | ||
== '''Catalytic Triad''' == | == '''Catalytic Triad''' == | ||
Revision as of 21:46, 15 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Contents |
Enoyl-ACP Reductase InhA from Mycobacterium tuberculosis
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Bell, A.F. et al.(2007). Evidence from Raman Spectroscopy That InhA , the Mycobacterial Enoyl Reductase, Modulates the Conformation of the NADH Cofactor to Promote Catalysis. Journal of the American Chemical Society, 129, 6425-6431. DOI: 10.1021/ja068219m
- ↑ 2.0 2.1 2.2 2.3 2.4 Bhatt, A. et al. (2007). The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Journal of Molecular Microbiology, 64(6), 1442-1454. PMID: 17555433 DOI: 10.1111/j.1365-2958.2007.05761.x
- ↑ Marrakchi, Hedia, et al. (2000). InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Journal of Microbiology, 146, 289-296. PMID: 10708367
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 Rozwarski, D.A. et al. (1999). Crystal Structure of the Mycobacterium tuberculosis Enoyl-ACP Reductase, InhA, in Complex with NAD+ and a C16 Fatty Acyl Substrate. Journal of Biological Chemistry, 274(22), 15582-15589. PMID: 10336454 DOI: 10.1074/jbc.274.22.15582
- ↑ Li, H.J. “et al.” (2014). A Structural and Energetic Model for the Slow-Onset Inhibition of the “Mycobacterium tuberculosis“ Enoyl-ACP Reductase InhA. “ACS Chemical Biology 9,” 986-993. PMID: 24527857
- ↑ Kruh, N. “et al.” (2007). Probing mechanisms of resistance to the tuberculosis drug isoniazid: Conformational changes caused by inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis. Protein Sci, 16(8), 1617-1627. PMID: 17600151
- ↑ 7.0 7.1 7.2 Vilchèze, C. et al. (2011). Novel Inhibitors of InhA Efficiently Kill Mycobacterium tuberculosis under Aerobic and Anaerobic Conditions. Antimicrobial Agents and Chemotherapy, 55(8), 3889-3898. DOI: 10.1128/AAC.00266-11
Student Contributors
- Arielle Russell
- Mackenzie A. Smith
Similar Proteopedia Pages
Enoyl-Acyl-Carrier Protein Reductase
Additional 3D Structures of Enoyl-ACP Reductase InhA
3oew, 2x22, 2x23, 1eny, 1enz, 4dqu, 4dre - MtENR+NAD; 3of2, 4dti - MtENR(mutant)+NAD; 2pr2, 2idz, 2h9i - MtENR+INH-NAPD; 2aq8 - MtENR+NADH; 2aqh, 2aqi, 2aqk, 3oey - MtENR(mutant)+NADH; 2ntj - MtENR+PTH-NAD; 2ie0, 2ieb, 2nv6, 1zid - MtENR(mutant)+INH-NAPD; 3fne, 3fnf, 3fng, 3fnh, 2b35, 1p45 - MtENR+NAD+TCI; 2b36, 2b37, 4ohu, 4oim, 4oyr - MtENR+NAD+phenoxyphenol derivative; 2nsd - MtENR+NAD+piperidine derivative; 2h7l, 2h7m, 2h7n, 2h7p, 4u0j, 4tzt, 4tzk, 4trj, 4u0k - MtENR+NAD+pyrrolidine derivative; 4cod, 4bqp, 4bqr, 4bge, 4bii, 4oxk, 4oxn, 4oxy, 4r9r, 4r9s - MtENR+NAD + inhibitor; 4bgi - MtENR (mutant)+NAD+inhibitor; 1p44 - MtENR+NAD+indole derivative; 1bvr - MtENR+NAD+fatty-acyl substrate