We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1075
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
Through specific binding factors, an EspG binds to its PE-PPE ligand to be secreted through the ESAT pathway. Though the ESAT-6 secretion system is poorly understood, it is known that PE-PPE proteins and EspG proteins influence virulence and pathogenicity of the infection. | Through specific binding factors, an EspG binds to its PE-PPE ligand to be secreted through the ESAT pathway. Though the ESAT-6 secretion system is poorly understood, it is known that PE-PPE proteins and EspG proteins influence virulence and pathogenicity of the infection. | ||
| - | |||
| - | |||
== Excretion == | == Excretion == | ||
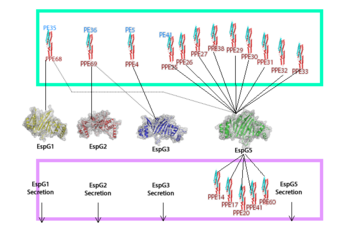
| - | EspG PE-PPE excretion is done through the ESX secretion pathway. Mycobacterium tuberculosis uses this ESX-1 secretion system to deliver virulence factors into host macrophage and monocyte white blood cells during the 'Mycobacterium tuberculosis' infection. Though the exact excreted protein is currently unknown, several different PE-PPE complexes and EspG complexes have been identified. In the image "Specificity of EspG Binding", it can be noted how the specificity of specific PE-PPE complexes only bind to certain EspG proteins. To exemplify this, notice the factors that influence | + | EspG PE-PPE excretion is done through the ESX secretion pathway. Mycobacterium tuberculosis uses this ESX-1 secretion system to deliver virulence factors into host macrophage and monocyte white blood cells during the 'Mycobacterium tuberculosis' infection. Though the exact excreted protein is currently unknown, several different PE-PPE complexes and EspG complexes have been identified. In the image "Specificity of EspG Binding", it can be noted how the specificity of specific PE-PPE complexes only bind to certain EspG proteins. To exemplify this, notice the factors that influence binding in EspG5 with PE25-PPE41 and see the contrast when EspG3 attempts PE25-PPE41. <ref>PMID:15973432</ref> |
[[Image:EspG_PPE-PE_secretion.fw_(2).png|350 px|left|thumb|[http://en.wikipedia.org/wiki/CFP-10 "Specificity of EspG Binding"]]] | [[Image:EspG_PPE-PE_secretion.fw_(2).png|350 px|left|thumb|[http://en.wikipedia.org/wiki/CFP-10 "Specificity of EspG Binding"]]] | ||
| Line 27: | Line 25: | ||
'''Residue Interactions:''' | '''Residue Interactions:''' | ||
| + | |||
<scene name='69/694242/Specific_contact_residues/1'>Random Loop, β2-β3 interactions</scene> | <scene name='69/694242/Specific_contact_residues/1'>Random Loop, β2-β3 interactions</scene> | ||
The random loop on the cigar shaped PE-PPE ligand binds to the β2-β3 sheets on this EspG protein. The residues on the random turn in the PE-PPE protein are key for the specificity to the EspG protein. Coils between PE-PPE proteins vary greatly and influence binding affinity. Combined with its β-2 & β-3 interactions on the EspG protein, the EspG protein is PE-PPE specific. These make up the bulk of residue interactions in the complex. An EspG protein will not bind to its non-corresponding PE-PPE complex. The residue hinderance image shows the hypothetical consequences of a non-corresponding EspG-PE-PPE bound complex. Here we see specific amino acid residue contact hinderance between the Asp36 and Gln172 on the EspG, with Asn122, His2, Phe3 and Met1 on the PE25-PPE41 complex. | The random loop on the cigar shaped PE-PPE ligand binds to the β2-β3 sheets on this EspG protein. The residues on the random turn in the PE-PPE protein are key for the specificity to the EspG protein. Coils between PE-PPE proteins vary greatly and influence binding affinity. Combined with its β-2 & β-3 interactions on the EspG protein, the EspG protein is PE-PPE specific. These make up the bulk of residue interactions in the complex. An EspG protein will not bind to its non-corresponding PE-PPE complex. The residue hinderance image shows the hypothetical consequences of a non-corresponding EspG-PE-PPE bound complex. Here we see specific amino acid residue contact hinderance between the Asp36 and Gln172 on the EspG, with Asn122, His2, Phe3 and Met1 on the PE25-PPE41 complex. | ||
| Line 35: | Line 34: | ||
'''Concavity:''' | '''Concavity:''' | ||
| - | <scene name='69/694242/Espg5_surface_with_pe-ppe/1'>EspG5 Concavity with PE-PPE bound</scene> | + | <scene name='69/694242/Espg5_surface_with_pe-ppe/1'>EspG5 Concavity with PE-PPE bound</scene> |
| - | + | ||
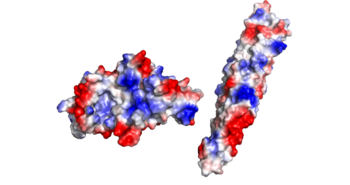
The concave region on the C-terminal half of the EspG protein facilitates binding of the tip of the cigar shaped PE-PPE. The tight bind between the protein and ligand works with the hydrophobic effect to increase binding affinity for the specific EspG-PE-PPE complex. | The concave region on the C-terminal half of the EspG protein facilitates binding of the tip of the cigar shaped PE-PPE. The tight bind between the protein and ligand works with the hydrophobic effect to increase binding affinity for the specific EspG-PE-PPE complex. | ||
| Line 42: | Line 40: | ||
'''Binding Pocket Residues ''' | '''Binding Pocket Residues ''' | ||
| - | EspG5 can bind to PE25-PPE41 due to a hand full of amino acid interactions. Most notably we have a Pro51 on alpha-2 helix of the EspG5 protein. Also we have various contact residues on the random turn that interact with the β2-β3 subunit, particularly the Glu127 of the random turn on the PE-PPE ligand. There are a few hydrophobic residues on the PE-PPE protein involved with binding affinity, the specific residues are Ala124, Leu125, Trp143, Gly147. | + | EspG5 can bind to PE25-PPE41 due to a hand full of amino acid interactions. Most notably we have a Pro51 on alpha-2 helix of the EspG5 protein. Also we have various contact residues on the random turn that interact with the β2-β3 subunit, particularly the Glu127 of the random turn on the PE-PPE ligand. There are a few hydrophobic residues on the PE-PPE protein involved with binding affinity, the specific residues are Ala124, Leu125, Trp143, Gly147. These hydrophobic regions are buried after the binding of PE-PPE to EspG, which exemplifies to the hydrophobic effect due to the increase in entropy of water. |
<scene name='69/694242/Hydrophobic_ppe_residues/1'>Hydrophobic Residues</scene> | <scene name='69/694242/Hydrophobic_ppe_residues/1'>Hydrophobic Residues</scene> | ||
| Line 65: | Line 63: | ||
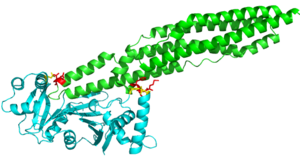
<scene name='69/694242/Espg3_differences/1'>EspG3 Compared to EspG5</scene> | <scene name='69/694242/Espg3_differences/1'>EspG3 Compared to EspG5</scene> | ||
| - | Here are the key differences found on the tertiary structure of EspG3 with EspG5. The highlighted alpha helix shows a difference in length between EspG3 and EspG5. This difference in length contributes to the steric hinderance when binding to a PE-PPE ligand. The random loop highlighted toward the bottom of this protein varies in length between EspG proteins, this influences binding. The small random turn highlighted shows inconceivable difference to the EspG5 protein. The β-2, β-3 sequence similarity between EspG5 and EspG3 is very low. | + | Here are the key differences found on the tertiary structure of EspG3 (yellow) with EspG5(blue). The highlighted alpha helix shows a difference in length between EspG3 and EspG5. This difference in length contributes to the steric hinderance when binding to a PE-PPE ligand. The random loop highlighted toward the bottom of this protein varies in length between EspG proteins, this influences binding. The small random turn highlighted shows inconceivable difference to the EspG5 protein. The β-2, β-3 sequence similarity between EspG5 and EspG3 is very low. |
Revision as of 00:51, 16 April 2015
| |||||||||||
References
- ↑ Ekiert DC, Cox JS. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14758-63. doi:, 10.1073/pnas.1409345111. Epub 2014 Oct 1. PMID:25275011 doi:http://dx.doi.org/10.1073/pnas.1409345111
- ↑ Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 2005 Jul 20;24(14):2491-8. Epub 2005 Jun 23. PMID:15973432
- ↑ Ekiert DC, Cox JS. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14758-63. doi:, 10.1073/pnas.1409345111. Epub 2014 Oct 1. PMID:25275011 doi:http://dx.doi.org/10.1073/pnas.1409345111
Similar Pages
Student Contributors
- Mark Meredith
- Jonathan Golliher