Introduction
EspG is a key secretion protein involved with the virulence of Mycobacterium tuberculosis. The specificity of EspG binding affinity to its specific PE-PPE ligand has many contributing factors. The four different EspG proteins found in Mycobacterium tuberculosis have different characteristics that influence binding, where EspG5 binds to the most PE-PPE proteins. Not all EspG proteins bind to the same ligand; specific interactions from specific residue interactions, electrostatics, steric hinderance and concavity of the EspG binding pocket influence binding. The EspG PE-PPE complex is to be excreted in the ESAT-6 pathway, this pathway is an attractive target for inducing apoptosis in Mtb, this makes it a good drug target.
Binding Specificity of EspG5 to PE25-PPE41 Proteins in Mycobacterium tuberculosis
General Structure and Function
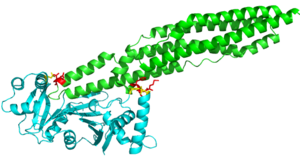
The crystal structure of EspG shows the protein in solution. As a monomeric protein, this binds to its ligand with high specificity. The EspG3 protein shown has a mass of 33.7kD [1] . The proteins beta sheets make up the backbone of the protein (yellow). A key beta sheet region on the will vary between EspG proteins to influence what the random loop on the PE-PPE protein will interact with. This backbone includes the key residues that interact with the random coil of the ligand. This is surrounded by 8 alpha helices (red) which add to the structure of the protein. One key will vary per EspG protein to stericly limit binding to PE-PPE ligand. The random coil (green) connects the beta sheets and helices together, where the long variations can impact binding to the EspG's ligand.
Through specific binding factors, an EspG binds to its PE-PPE ligand to be secreted through the ESAT pathway. Though the ESAT-6 secretion system is poorly understood, it is known that PE-PPE proteins and EspG proteins influence virulence and pathogenicity of the infection.
Excretion
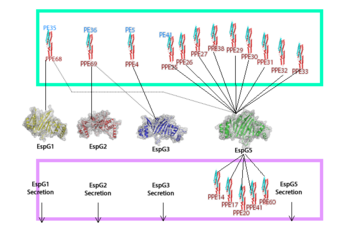
EspG PE-PPE excretion is done through the ESX secretion pathway. Mycobacterium tuberculosis uses this ESX-1 secretion system to deliver virulence factors into host macrophage and monocyte white blood cells during the 'Mycobacterium tuberculosis' infection. Though the exact excreted protein is currently unknown, several different PE-PPE complexes and EspG complexes have been identified. In the image "Specificity of EspG Binding", it can be noted how the specificity of specific PE-PPE complexes only bind to certain EspG proteins. To exemplify this, notice the factors that influence binding in EspG5 with PE25-PPE41 and see the contrast when EspG3 attempts PE25-PPE41. [2]
Binding
The EspG-PE-PPE binding is highly specific. The specific pair we were looking at was the EspG5-PE25-PPE41 complex. A variety of binding factors influence the EspG that will bind to a specific PE-PPE. The secretion pathway carried out needs the coupled protein.
Residue Interactions:
The random loop on the cigar shaped PE-PPE ligand binds to the β2-β3 sheets on this EspG protein. The residues on the random turn in the PE-PPE protein are key for the specificity to the EspG protein. Coils between PE-PPE proteins vary greatly and influence binding affinity. Combined with its β-2 & β-3 interactions on the EspG protein, the EspG protein is PE-PPE specific. These make up the bulk of residue interactions in the complex. An EspG protein will not bind to its non-corresponding PE-PPE complex. The residue hinderance image shows the hypothetical consequences of a non-corresponding EspG-PE-PPE bound complex. Here we see specific amino acid residue contact hinderance between the Asp36 and Gln172 on the EspG, with Asn122, His2, Phe3 and Met1 on the PE25-PPE41 complex.
Concavity:
The concave region on the C-terminal half of the EspG protein facilitates binding of the tip of the cigar shaped PE-PPE. The tight bind between the protein and ligand works with the hydrophobic effect to increase binding affinity for the specific EspG-PE-PPE complex. These hydrophobic regions are buried after the binding of PE-PPE to EspG, which exemplifies to the hydrophobic effect due to the increase in entropy of water.
Binding Pocket Residues
EspG5 can bind to PE25-PPE41 due to a hand full of amino acid interactions. Most notably we have a Pro51 on α-2 helix of the EspG5 protein. Also we have various contact residues on the random turn that interact with the β2-β3 subunit. Particularly between the Glu127 of the random turn on the PE-PPE ligand interacts with the Glu256 on the EspG protein. There are a few hydrophobic residues on the PE-PPE protein involved with binding affinity, the specific residues are .
Electrostatics:
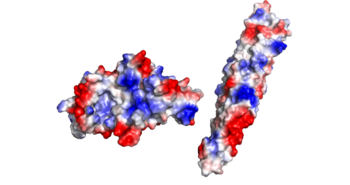
There is an overall negative charge on the PE-PPE complex, and the binding tip is only partially negative. On the EspG5 protein, the binding pocket is partially positive, which aids coupling of the EspG & PE-PPE complex. The other EspG proteins found in 'Mycobacterium tuberculosis' have different electrostatic pocket charges which prevent binding of the PE25-PPE41 ligand.
Structural highlights
Here are the key differences found on the tertiary structure of EspG3 (yellow) with EspG5(blue). The highlighted alpha helix shows a difference in length between EspG3 and EspG5. This difference in length contributes to the steric hinderance when binding to a PE-PPE ligand. The random loop highlighted toward the bottom of this protein varies in length between EspG proteins, this influences binding. The small random turn highlighted shows inconceivable difference to the EspG5 protein. The β-2, β-3 sequence similarity between EspG5 and EspG3 is very low.
Relevance
The binding affinity between EspG and PE-PPE ligands is needed for excretion into the ESAT-6 pathway in Mycobacterium tuberculosis. This pathway is of popular study of Mtb because it is linked to the virulence of the virus. Hindering this pathway has been found to induce apoptosis in cells infested with Mtb.
Publication Abstract from PubMed
Nearly 10% of the coding capacity of the Mycobacterium tuberculosis genome is devoted to two highly expanded and enigmatic protein families called PE and PPE, some of which are important virulence/immunogenicity factors and are secreted during infection via a unique alternative secretory system termed "type VII." How PE-PPE proteins function during infection and how they are translocated to the bacterial surface through the five distinct type VII secretion systems [ESAT-6 secretion system (ESX)] of M. tuberculosis is poorly understood. Here, we report the crystal structure of a PE-PPE heterodimer bound to ESX secretion-associated protein G (EspG), which adopts a novel fold. This PE-PPE-EspG complex, along with structures of two additional EspGs, suggests that EspG acts as an adaptor that recognizes specific PE-PPE protein complexes via extensive interactions with PPE domains, and delivers them to ESX machinery for secretion. Surprisingly, secretion of most PE-PPE proteins in M. tuberculosis is likely mediated by EspG from the ESX-5 system, underscoring the importance of ESX-5 in mycobacterial pathogenesis. Moreover, our results indicate that PE-PPE domains function as cis-acting targeting sequences that are read out by EspGs, revealing the molecular specificity for secretion through distinct ESX pathways.
Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion.,Ekiert DC, Cox JS Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14758-63. doi:, 10.1073/pnas.1409345111. Epub 2014 Oct 1. PMID:25275011[3]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.