We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1066
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
<StructureSection load='3r44' size='340' side='right' caption='Very Long Chain Fatty Acyl CoA Synthetase (FadD13)' scene='69/694232/Opening_scene/1'> | <StructureSection load='3r44' size='340' side='right' caption='Very Long Chain Fatty Acyl CoA Synthetase (FadD13)' scene='69/694232/Opening_scene/1'> | ||
| - | |||
= Introduction = | = Introduction = | ||
| Line 10: | Line 9: | ||
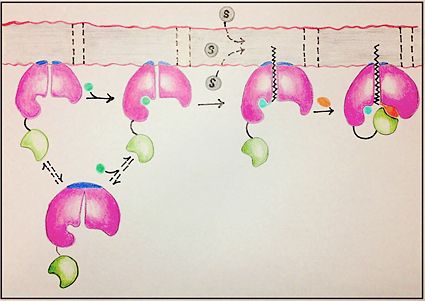
There are four main groups of FadD enzymes categorized on their ability to accommodate different length substrates: short (C2-C4), medium (C4-C12), long (C12-C22), and very long (C22-C26).<ref name="Our Paper"/> Most FadD class proteins exist as integral membrane proteins, involved in both the activation of fatty acids and other hydrophobic substrates in addition to the transport of these lipids into the cell. However, substrates longer than the enzyme itself, like these very-long-chain fatty acids, pose an interesting structural dilemma to the enzyme. FadD13 differs from typical integral membrane fatty acyl-CoA synthetases in that FadD13 exists as a [https://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral membrane protein]. This key feature provides a mechanistic basis for FadD13’s activation and transport of fatty acids of length C24-C26 through the two step addition of [http://en.wikipedia.org/wiki/Coenzyme_A Coenzyme A](Figure 1).<ref name="Our Paper">PMID: 22560731</ref> | There are four main groups of FadD enzymes categorized on their ability to accommodate different length substrates: short (C2-C4), medium (C4-C12), long (C12-C22), and very long (C22-C26).<ref name="Our Paper"/> Most FadD class proteins exist as integral membrane proteins, involved in both the activation of fatty acids and other hydrophobic substrates in addition to the transport of these lipids into the cell. However, substrates longer than the enzyme itself, like these very-long-chain fatty acids, pose an interesting structural dilemma to the enzyme. FadD13 differs from typical integral membrane fatty acyl-CoA synthetases in that FadD13 exists as a [https://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral membrane protein]. This key feature provides a mechanistic basis for FadD13’s activation and transport of fatty acids of length C24-C26 through the two step addition of [http://en.wikipedia.org/wiki/Coenzyme_A Coenzyme A](Figure 1).<ref name="Our Paper">PMID: 22560731</ref> | ||
| - | |||
= Mechanism = | = Mechanism = | ||
[[Image:FadD13 edited image.jpg|425 px|left|thumb|Figure 1: Mechanism for the activation of fatty acids (C24-C26) by FadD13. The N terminal domain (pink) is embedded in the membrane with the arginine rich lid-loop (dark blue), while the flexile linker (black) connects the C terminal domain (green) to the rest of the enzyme. Activation requires the binding of ATP (blue) which induces structural changes that promote the binding of the fatty acid chain. Formation of an acyl-adenylate intermediate induces a 140° rotation of the C terminal domain and the binding of CoA (orange). ]] | [[Image:FadD13 edited image.jpg|425 px|left|thumb|Figure 1: Mechanism for the activation of fatty acids (C24-C26) by FadD13. The N terminal domain (pink) is embedded in the membrane with the arginine rich lid-loop (dark blue), while the flexile linker (black) connects the C terminal domain (green) to the rest of the enzyme. Activation requires the binding of ATP (blue) which induces structural changes that promote the binding of the fatty acid chain. Formation of an acyl-adenylate intermediate induces a 140° rotation of the C terminal domain and the binding of CoA (orange). ]] | ||
| - | |||
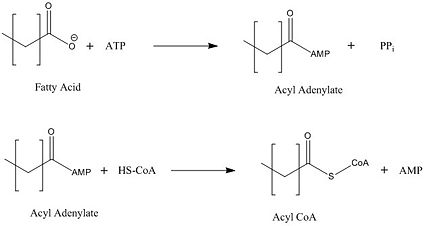
[[Image:acyl coa synthetase.jpg|425 px|right|thumb|Figure 2: Representation of the two-step reaction catalyzed by FadD13]] | [[Image:acyl coa synthetase.jpg|425 px|right|thumb|Figure 2: Representation of the two-step reaction catalyzed by FadD13]] | ||
| Line 20: | Line 17: | ||
== General mechanism for the activation of fatty acids == | == General mechanism for the activation of fatty acids == | ||
FadD13 represents the first Fatty Acyl-CoA Synthetase of its kind to display biphasic kinetics.<ref name="residue paper"/> FadD13 first activates the fatty acid through a reaction with ATP to form an acyl adenylate intermediate and subsequently releases a pyrophosphate. Following a conformational change of the enzyme upon the binding of ATP, coenzyme A is able to bind to its active site and react with the acyl adenylate intermediate forming the [http://en.wikipedia.org/wiki/Acyl-CoA acyl CoA] product (Figure 2). These activated fatty acyl-CoA thioesters have then been demonstrated to be important for the synthesis of triacyglycerols and phospholipids in the membrane of ''Mycobacterium tuberculosis''. <ref name="residue paper"/> | FadD13 represents the first Fatty Acyl-CoA Synthetase of its kind to display biphasic kinetics.<ref name="residue paper"/> FadD13 first activates the fatty acid through a reaction with ATP to form an acyl adenylate intermediate and subsequently releases a pyrophosphate. Following a conformational change of the enzyme upon the binding of ATP, coenzyme A is able to bind to its active site and react with the acyl adenylate intermediate forming the [http://en.wikipedia.org/wiki/Acyl-CoA acyl CoA] product (Figure 2). These activated fatty acyl-CoA thioesters have then been demonstrated to be important for the synthesis of triacyglycerols and phospholipids in the membrane of ''Mycobacterium tuberculosis''. <ref name="residue paper"/> | ||
| - | |||
== Structural basis for housing lipid substrates longer than the enzyme == | == Structural basis for housing lipid substrates longer than the enzyme == | ||
| Line 42: | Line 38: | ||
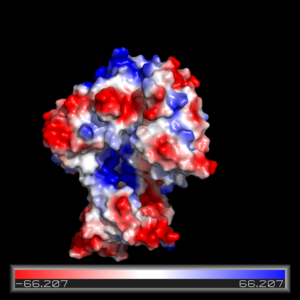
The FadD13 active site is composed of positively charged regions which account for the attraction and binding of hydrophobic substrates to this region (Figure 3). The active site on FadD13 is composed of two conserved regions, one of which serves as the binding site for ATP and the other for CoA. The adenine of ATP is bound to a group of <scene name='69/694232/Adenine_binding_group/2'>six amino acids (300-305)</scene> that is structurally identically to other acyl-CoA synthetases. <ref name="Our Paper"/> | The FadD13 active site is composed of positively charged regions which account for the attraction and binding of hydrophobic substrates to this region (Figure 3). The active site on FadD13 is composed of two conserved regions, one of which serves as the binding site for ATP and the other for CoA. The adenine of ATP is bound to a group of <scene name='69/694232/Adenine_binding_group/2'>six amino acids (300-305)</scene> that is structurally identically to other acyl-CoA synthetases. <ref name="Our Paper"/> | ||
| - | Mutation of the highly conserved residue in the C-terminal region, <scene name='69/694233/Lys_487/2'>Lysine 487</scene>, resulted in a 95% loss of function of FadD13 and is thought to be involved in the orientation of the substrates to form the adenylate intermediate.<ref name="residue paper">PMID: 20027301</ref> Additionally, Serine 404 was hypothesized to be involved in the binding of Coenzyme A which may only occur once this region incurs a 140 degree rotational change after the initial binding of ATP.<ref name="Our Paper"/><ref name="residue paper"/> | + | Mutation of the highly conserved residue in the C-terminal region, <scene name='69/694233/Lys_487/2'>Lysine 487</scene>, resulted in a 95% loss of function of FadD13 and is thought to be involved in the orientation of the substrates to form the adenylate intermediate.<ref name="residue paper">PMID: 20027301</ref> Additionally, <scene name='69/694233/Ser_404/1'>Serine 404</scene> was hypothesized to be involved in the binding of Coenzyme A which may only occur once this region incurs a 140 degree rotational change after the initial binding of ATP.<ref name="Our Paper"/><ref name="residue paper"/> |
| - | + | ||
= Disease = | = Disease = | ||
Revision as of 17:50, 17 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Molina DM, Hogbom M. The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme. Structure. 2012 May 2. PMID:22560731 doi:10.1016/j.str.2012.03.012
- ↑ Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis--a potential target for the development of antitubercular drugs. J Mol Model. 2011 Feb;17(2):301-13. doi: 10.1007/s00894-010-0727-3. Epub 2010 May, 8. PMID:20454815 doi:http://dx.doi.org/10.1007/s00894-010-0727-3
- ↑ 3.0 3.1 3.2 3.3 3.4 Khare G, Gupta V, Gupta RK, Gupta R, Bhat R, Tyagi AK. Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA Synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One. 2009 Dec 21;4(12):e8387. doi: 10.1371/journal.pone.0008387. PMID:20027301 doi:10.1371/journal.pone.0008387
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Jatana N, Jangid S, Khare G, Tyagi AK, Latha N. Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis--a potential target for the development of antitubercular drugs. J Mol Model. 2011 Feb;17(2):301-13. doi: 10.1007/s00894-010-0727-3. Epub 2010 May, 8. PMID:20454815 doi:http://dx.doi.org/10.1007/s00894-010-0727-3
- ↑ 5.0 5.1 5.2 Schroeder EK, de Souza N, Santos DS, Blanchard JS, Basso LA. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis. Curr Pharm Biotechnol. 2002 Sep;3(3):197-225. PMID:12164478
External Resources
Lipids Wikipedia page
Tuberculosis Wikipedia page
Mycobacterium tuberculosis Wikipedia page
Coenzyme A Wikipedia page
Acyl CoA Wikipedia Page
Mycolic Acid Wikipedia page
Peripheral Membrane Protein Wikipedia page
Ethionamide Wikipedia page
Isoniazid Wikipedia page
Thiocarlide Wikipedia page
Pyrazinamide Wikipedia page