We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
==Future Work== | ==Future Work== | ||
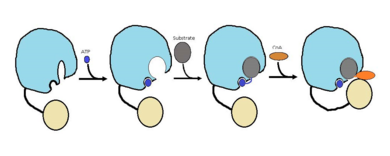
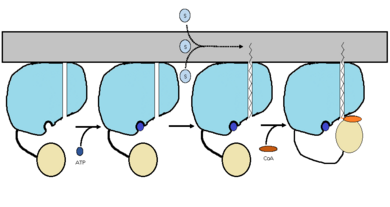
| - | Currently there is no crystal structure available for FadD13 complexes with lipids. Therefore the the mechanism that researchers proposed in (figure 2) is just based on the biochemical and structural information that they have been able to gather. The reason for the lack of a crystal structure for FadD13 complexes with ligands is due to the need for membrane interaction. The membrane interaction is required to induece a substrate binding conformation of the hydrophobic channel. In the future, researchers would like to develop a crystal structure of FadD13 peripherally bound to the membrane to gain a better understanding of how substrates bind. Despite the lack of a crystal structure, the proposed mechanism that researches developed (figure 2) could help synthesize inhibitors and locate certain locations for drug inhibitors to bind. | + | Currently there is no crystal structure available for FadD13 complexes with lipids. Therefore the the mechanism that researchers proposed in (figure 2) is just based on the biochemical and structural information that they have been able to gather. The reason for the lack of a crystal structure for FadD13 complexes with ligands is due to the need for membrane interaction. The membrane interaction is required to induece a substrate binding conformation of the hydrophobic channel. In the future, researchers would like to develop a crystal structure of FadD13 peripherally bound to the membrane to gain a better understanding of how substrates bind. Despite the lack of a crystal structure, the proposed mechanism that researches developed (figure 2) could help synthesize inhibitors and locate certain locations for drug inhibitors to bind.<ref name="Anderson 2012"/> |
==Relevant Pages== | ==Relevant Pages== | ||
Revision as of 19:03, 21 April 2015
FadD13
| |||||||||||
References
- ↑ Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 2007 Dec;48(12):2736-50. Epub 2007 Aug 30. PMID:17762044 doi:http://dx.doi.org/M700378-JLR200
- ↑ Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW. Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A. J Mol Biol. 2009 May 22;388(5):997-1008. Epub 2009 Apr 1. PMID:19345228 doi:10.1016/j.jmb.2009.03.064
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Molina DM, Hogbom M. The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme. Structure. 2012 May 2. PMID:22560731 doi:10.1016/j.str.2012.03.012
- ↑ 4.0 4.1 4.2 Khare G, Gupta V, Gupta RK, Gupta R, Bhat R, Tyagi AK. Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA Synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One. 2009 Dec 21;4(12):e8387. doi: 10.1371/journal.pone.0008387. PMID:20027301 doi:10.1371/journal.pone.0008387